Marco A. S. Netto
Context-aware Execution Migration Tool for Data Science Jupyter Notebooks on Hybrid Clouds
Jul 01, 2021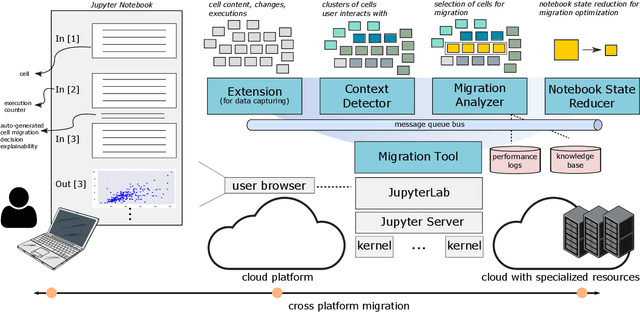
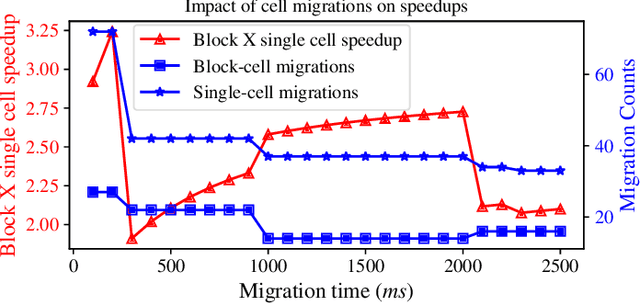
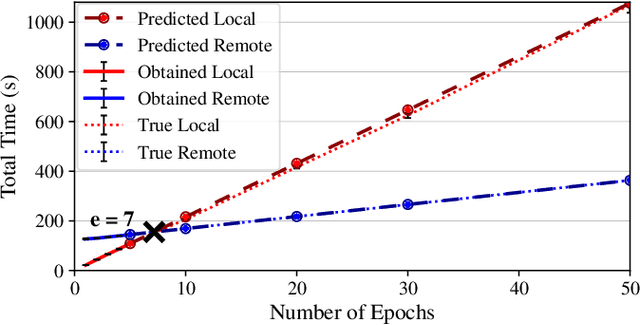
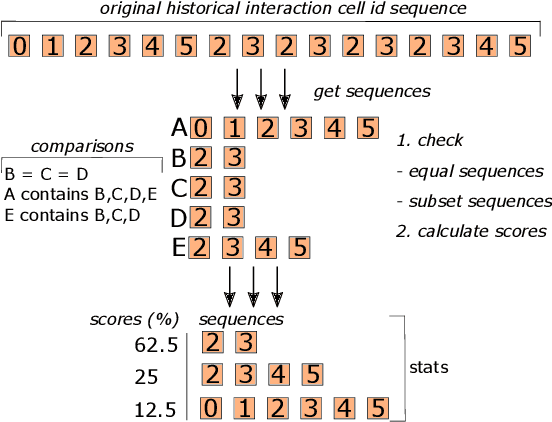
Abstract:Interactive computing notebooks, such as Jupyter notebooks, have become a popular tool for developing and improving data-driven models. Such notebooks tend to be executed either in the user's own machine or in a cloud environment, having drawbacks and benefits in both approaches. This paper presents a solution developed as a Jupyter extension that automatically selects which cells, as well as in which scenarios, such cells should be migrated to a more suitable platform for execution. We describe how we reduce the execution state of the notebook to decrease migration time and we explore the knowledge of user interactivity patterns with the notebook to determine which blocks of cells should be migrated. Using notebooks from Earth science (remote sensing), image recognition, and hand written digit identification (machine learning), our experiments show notebook state reductions of up to 55x and migration decisions leading to performance gains of up to 3.25x when the user interactivity with the notebook is taken into consideration.
Workflow Provenance in the Lifecycle of Scientific Machine Learning
Sep 30, 2020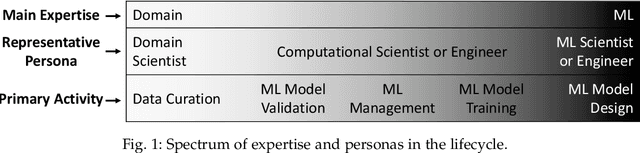
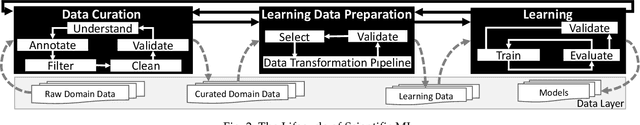
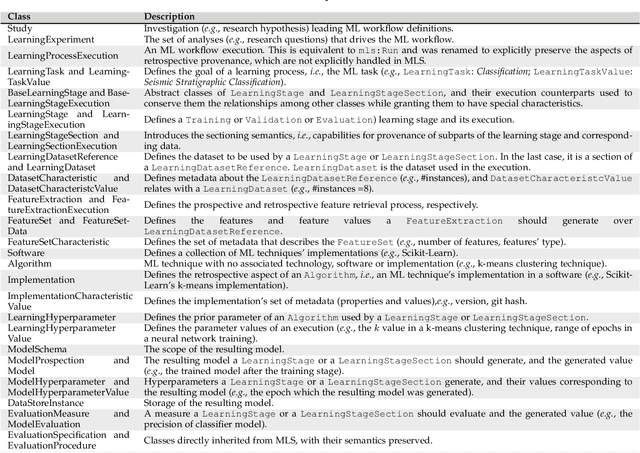
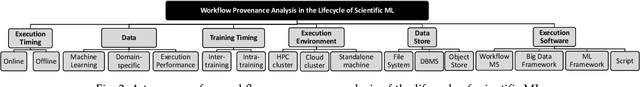
Abstract:Machine Learning (ML) has already fundamentally changed several businesses. More recently, it has also been profoundly impacting the computational science and engineering domains, like geoscience, climate science, and health science. In these domains, users need to perform comprehensive data analyses combining scientific data and ML models to provide for critical requirements, such as reproducibility, model explainability, and experiment data understanding. However, scientific ML is multidisciplinary, heterogeneous, and affected by the physical constraints of the domain, making such analyses even more challenging. In this work, we leverage workflow provenance techniques to build a holistic view to support the lifecycle of scientific ML. We contribute with (i) characterization of the lifecycle and taxonomy for data analyses; (ii) design principles to build this view, with a W3C PROV compliant data representation and a reference system architecture; and (iii) lessons learned after an evaluation in an Oil & Gas case using an HPC cluster with 393 nodes and 946 GPUs. The experiments show that the principles enable queries that integrate domain semantics with ML models while keeping low overhead (<1%), high scalability, and an order of magnitude of query acceleration under certain workloads against without our representation.
Provenance Data in the Machine Learning Lifecycle in Computational Science and Engineering
Oct 21, 2019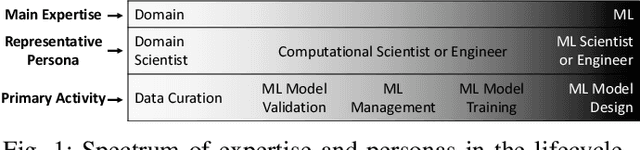
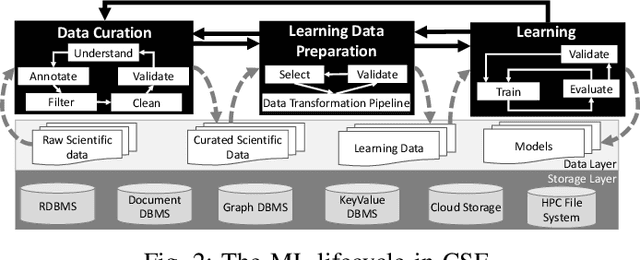
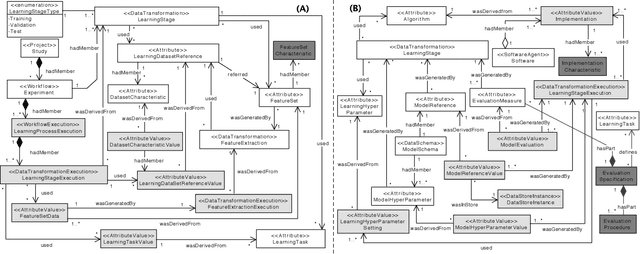
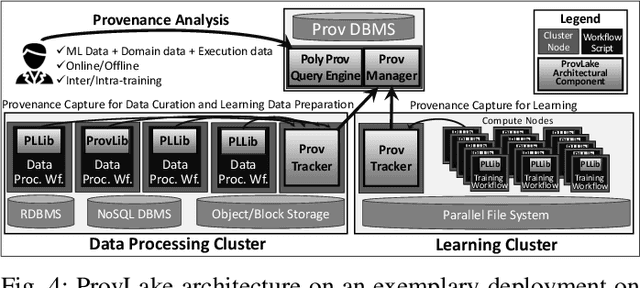
Abstract:Machine Learning (ML) has become essential in several industries. In Computational Science and Engineering (CSE), the complexity of the ML lifecycle comes from the large variety of data, scientists' expertise, tools, and workflows. If data are not tracked properly during the lifecycle, it becomes unfeasible to recreate a ML model from scratch or to explain to stakeholders how it was created. The main limitation of provenance tracking solutions is that they cannot cope with provenance capture and integration of domain and ML data processed in the multiple workflows in the lifecycle while keeping the provenance capture overhead low. To handle this problem, in this paper we contribute with a detailed characterization of provenance data in the ML lifecycle in CSE; a new provenance data representation, called PROV-ML, built on top of W3C PROV and ML Schema; and extensions to a system that tracks provenance from multiple workflows to address the characteristics of ML and CSE, and to allow for provenance queries with a standard vocabulary. We show a practical use in a real case in the Oil and Gas industry, along with its evaluation using 48 GPUs in parallel.
A Scalable Machine Learning System for Pre-Season Agriculture Yield Forecast
Oct 15, 2018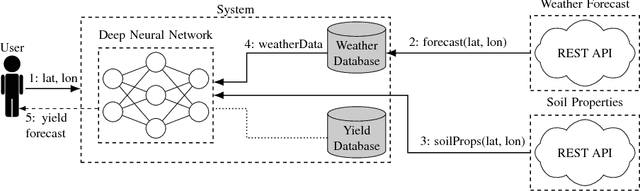
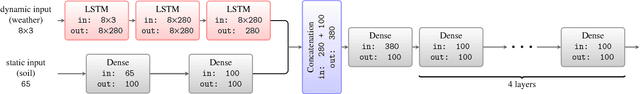

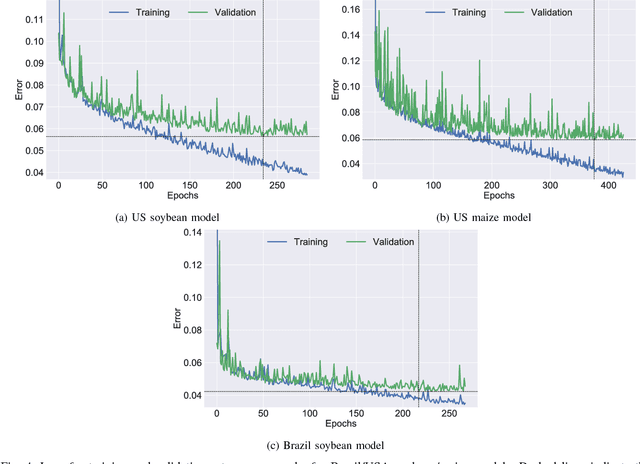
Abstract:Yield forecast is essential to agriculture stakeholders and can be obtained with the use of machine learning models and data coming from multiple sources. Most solutions for yield forecast rely on NDVI (Normalized Difference Vegetation Index) data, which is time-consuming to be acquired and processed. To bring scalability for yield forecast, in the present paper we describe a system that incorporates satellite-derived precipitation and soil properties datasets, seasonal climate forecasting data from physical models and other sources to produce a pre-season prediction of soybean/maize yield---with no need of NDVI data. This system provides significantly useful results by the exempting the need for high-resolution remote-sensing data and allowing farmers to prepare for adverse climate influence on the crop cycle. In our studies, we forecast the soybean and maize yields for Brazil and USA, which corresponded to 44% of the world's grain production in 2016. Results show the error metrics for soybean and maize yield forecasts are comparable to similar systems that only provide yield forecast information in the first weeks to months of the crop cycle.
DeepDownscale: a Deep Learning Strategy for High-Resolution Weather Forecast
Aug 15, 2018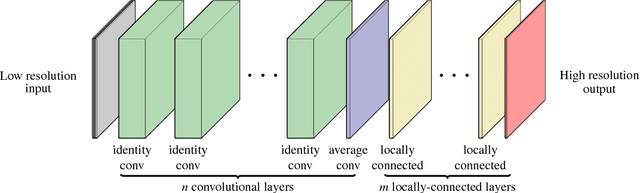
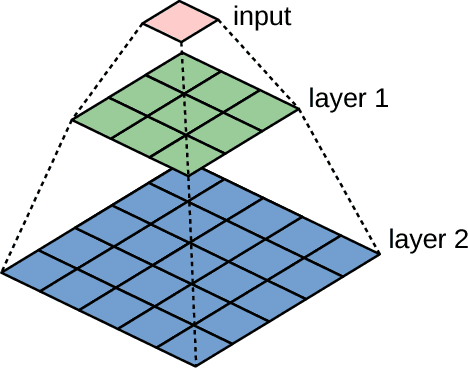
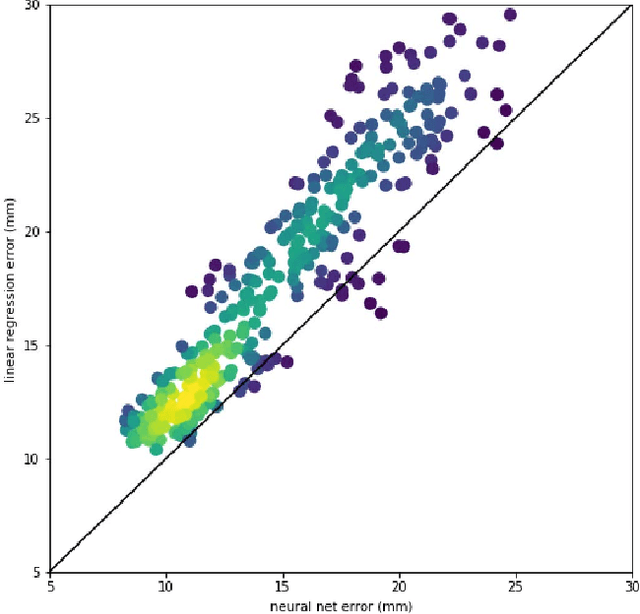
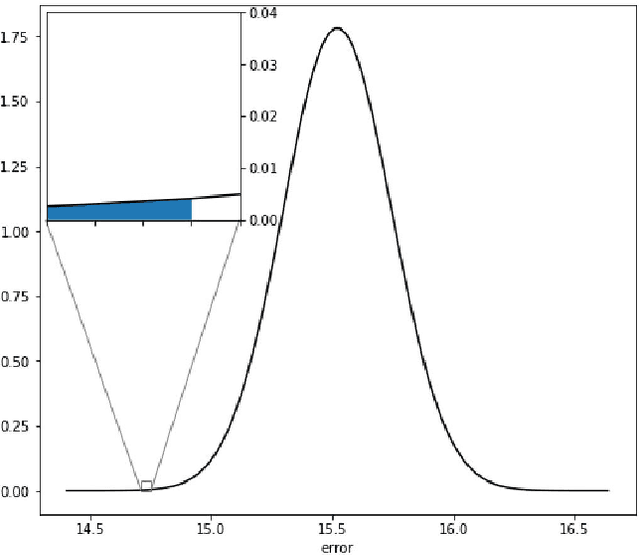
Abstract:Running high-resolution physical models is computationally expensive and essential for many disciplines. Agriculture, transportation, and energy are sectors that depend on high-resolution weather models, which typically consume many hours of large High Performance Computing (HPC) systems to deliver timely results. Many users cannot afford to run the desired resolution and are forced to use low resolution output. One simple solution is to interpolate results for visualization. It is also possible to combine an ensemble of low resolution models to obtain a better prediction. However, these approaches fail to capture the redundant information and patterns in the low-resolution input that could help improve the quality of prediction. In this paper, we propose and evaluate a strategy based on a deep neural network to learn a high-resolution representation from low-resolution predictions using weather forecast as a practical use case. We take a supervised learning approach, since obtaining labeled data can be done automatically. Our results show significant improvement when compared with standard practices and the strategy is still lightweight enough to run on modest computer systems.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge