Mahdi Ghorbani
Compressing Structured Tensor Algebra
Jul 18, 2024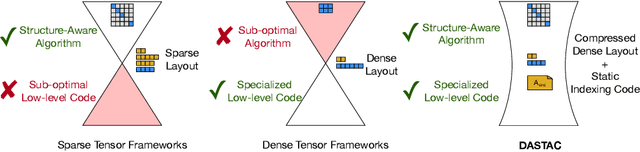
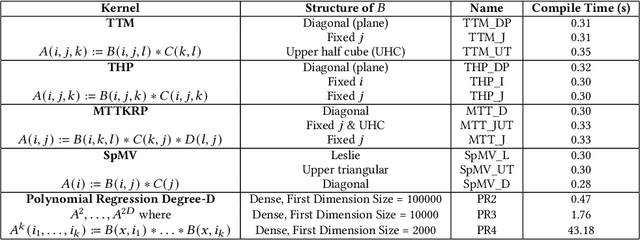


Abstract:Tensor algebra is a crucial component for data-intensive workloads such as machine learning and scientific computing. As the complexity of data grows, scientists often encounter a dilemma between the highly specialized dense tensor algebra and efficient structure-aware algorithms provided by sparse tensor algebra. In this paper, we introduce DASTAC, a framework to propagate the tensors's captured high-level structure down to low-level code generation by incorporating techniques such as automatic data layout compression, polyhedral analysis, and affine code generation. Our methodology reduces memory footprint by automatically detecting the best data layout, heavily benefits from polyhedral optimizations, leverages further optimizations, and enables parallelization through MLIR. Through extensive experimentation, we show that DASTAC achieves 1 to 2 orders of magnitude speedup over TACO, a state-of-the-art sparse tensor compiler, and StructTensor, a state-of-the-art structured tensor algebra compiler, with a significantly lower memory footprint.
GraphVAMPNet, using graph neural networks and variational approach to markov processes for dynamical modeling of biomolecules
Jan 12, 2022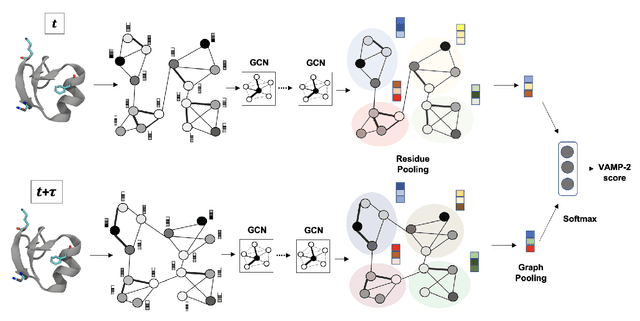

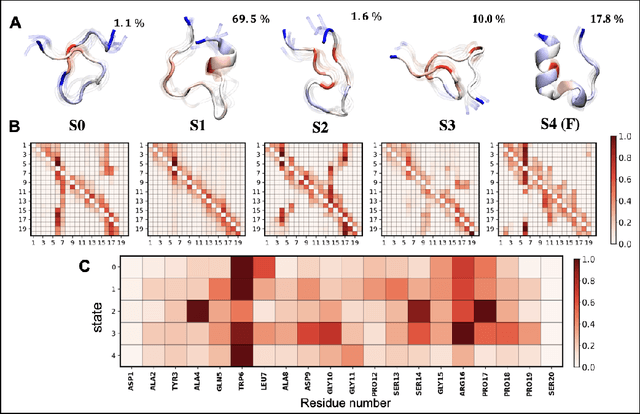
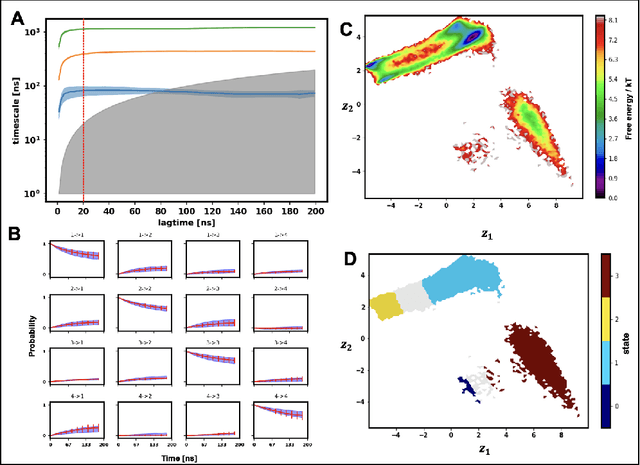
Abstract:Finding low dimensional representation of data from long-timescale trajectories of biomolecular processes such as protein-folding or ligand-receptor binding is of fundamental importance and kinetic models such as Markov modeling have proven useful in describing the kinetics of these systems. Recently, an unsupervised machine learning technique called VAMPNet was introduced to learn the low dimensional representation and linear dynamical model in an end-to-end manner. VAMPNet is based on variational approach to Markov processes (VAMP) and relies on neural networks to learn the coarse-grained dynamics. In this contribution, we combine VAMPNet and graph neural networks to generate an end-to-end framework to efficiently learn high-level dynamics and metastable states from the long-timescale molecular dynamics trajectories. This method bears the advantages of graph representation learning and uses graph message passing operations to generate an embedding for each datapoint which is used in the VAMPNet to generate a coarse-grained representation. This type of molecular representation results in a higher resolution and more interpretable Markov model than the standard VAMPNet enabling a more detailed kinetic study of the biomolecular processes. Our GraphVAMPNet approach is also enhanced with an attention mechanism to find the important residues for classification into different metastable states.
Fine-Tuning Data Structures for Analytical Query Processing
Dec 24, 2021
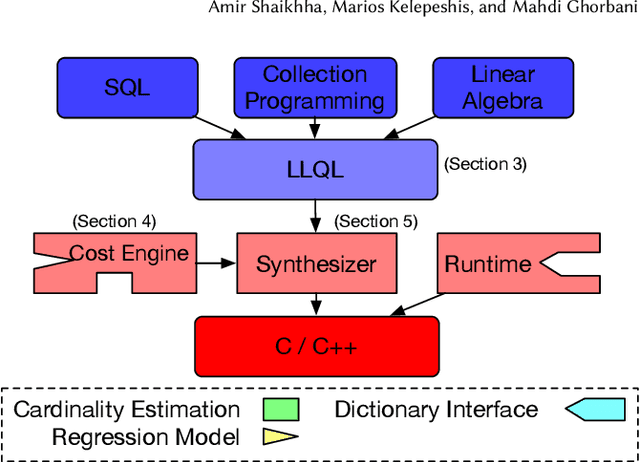
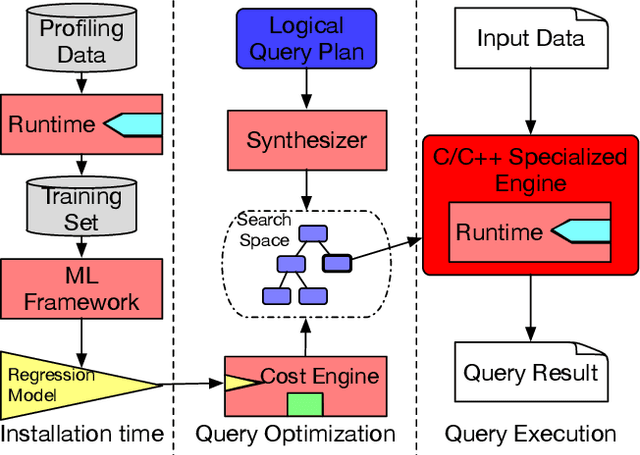

Abstract:We introduce a framework for automatically choosing data structures to support efficient computation of analytical workloads. Our contributions are twofold. First, we introduce a novel low-level intermediate language that can express the algorithms behind various query processing paradigms such as classical joins, groupjoin, and in-database machine learning engines. This language is designed around the notion of dictionaries, and allows for a more fine-grained choice of its low-level implementation. Second, the cost model for alternative implementations is automatically inferred by combining machine learning and program reasoning. The dictionary cost model is learned using a regression model trained over the profiling dataset of dictionary operations on a given hardware architecture. The program cost model is inferred using static program analysis. Our experimental results show the effectiveness of the trained cost model on micro benchmarks. Furthermore, we show that the performance of the code generated by our framework either outperforms or is on par with the state-of-the-art analytical query engines and a recent in-database machine learning framework.
Variational embedding of protein folding simulations using gaussian mixture variational autoencoders
Aug 27, 2021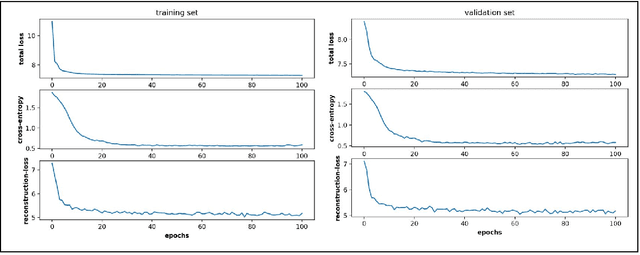
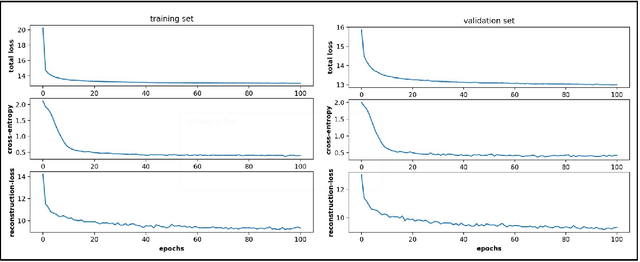


Abstract:Conformational sampling of biomolecules using molecular dynamics simulations often produces large amount of high dimensional data that makes it difficult to interpret using conventional analysis techniques. Dimensionality reduction methods are thus required to extract useful and relevant information. Here we devise a machine learning method, Gaussian mixture variational autoencoder (GMVAE) that can simultaneously perform dimensionality reduction and clustering of biomolecular conformations in an unsupervised way. We show that GMVAE can learn a reduced representation of the free energy landscape of protein folding with highly separated clusters that correspond to the metastable states during folding. Since GMVAE uses a mixture of Gaussians as the prior, it can directly acknowledge the multi-basin nature of protein folding free-energy landscape. To make the model end-to-end differentialble, we use a Gumbel-softmax distribution. We test the model on three long-timescale protein folding trajectories and show that GMVAE embedding resembles the folding funnel with folded states down the funnel and unfolded states outer in the funnel path. Additionally, we show that the latent space of GMVAE can be used for kinetic analysis and Markov state models built on this embedding produce folding and unfolding timescales that are in close agreement with other rigorous dynamical embeddings such as time independent component analysis (TICA).
Be Your Own Best Competitor! Multi-Branched Adversarial Knowledge Transfer
Oct 09, 2020



Abstract:Deep neural network architectures have attained remarkable improvements in scene understanding tasks. Utilizing an efficient model is one of the most important constraints for limited-resource devices. Recently, several compression methods have been proposed to diminish the heavy computational burden and memory consumption. Among them, the pruning and quantizing methods exhibit a critical drop in performances by compressing the model parameters. While the knowledge distillation methods improve the performance of compact models by focusing on training lightweight networks with the supervision of cumbersome networks. In the proposed method, the knowledge distillation has been performed within the network by constructing multiple branches over the primary stream of the model, known as the self-distillation method. Therefore, the ensemble of sub-neural network models has been proposed to transfer the knowledge among themselves with the knowledge distillation policies as well as an adversarial learning strategy. Hence, The proposed ensemble of sub-models is trained against a discriminator model adversarially. Besides, their knowledge is transferred within the ensemble by four different loss functions. The proposed method has been devoted to both lightweight image classification and encoder-decoder architectures to boost the performance of small and compact models without incurring extra computational overhead at the inference process. Extensive experimental results on the main challenging datasets show that the proposed network outperforms the primary model in terms of accuracy at the same number of parameters and computational cost. The obtained results show that the proposed model has achieved significant improvement over earlier ideas of self-distillation methods. The effectiveness of the proposed models has also been illustrated in the encoder-decoder model.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge