Lukas Drees
Climplicit: Climatic Implicit Embeddings for Global Ecological Tasks
Apr 07, 2025Abstract:Deep learning on climatic data holds potential for macroecological applications. However, its adoption remains limited among scientists outside the deep learning community due to storage, compute, and technical expertise barriers. To address this, we introduce Climplicit, a spatio-temporal geolocation encoder pretrained to generate implicit climatic representations anywhere on Earth. By bypassing the need to download raw climatic rasters and train feature extractors, our model uses x1000 fewer disk space and significantly reduces computational needs for downstream tasks. We evaluate our Climplicit embeddings on biomes classification, species distribution modeling, and plant trait regression. We find that linear probing our Climplicit embeddings consistently performs better or on par with training a model from scratch on downstream tasks and overall better than alternative geolocation encoding models.
Data-driven Crop Growth Simulation on Time-varying Generated Images using Multi-conditional Generative Adversarial Networks
Dec 06, 2023Abstract:Image-based crop growth modeling can substantially contribute to precision agriculture by revealing spatial crop development over time, which allows an early and location-specific estimation of relevant future plant traits, such as leaf area or biomass. A prerequisite for realistic and sharp crop image generation is the integration of multiple growth-influencing conditions in a model, such as an image of an initial growth stage, the associated growth time, and further information about the field treatment. We present a two-stage framework consisting first of an image prediction model and second of a growth estimation model, which both are independently trained. The image prediction model is a conditional Wasserstein generative adversarial network (CWGAN). In the generator of this model, conditional batch normalization (CBN) is used to integrate different conditions along with the input image. This allows the model to generate time-varying artificial images dependent on multiple influencing factors of different kinds. These images are used by the second part of the framework for plant phenotyping by deriving plant-specific traits and comparing them with those of non-artificial (real) reference images. For various crop datasets, the framework allows realistic, sharp image predictions with a slight loss of quality from short-term to long-term predictions. Simulations of varying growth-influencing conditions performed with the trained framework provide valuable insights into how such factors relate to crop appearances, which is particularly useful in complex, less explored crop mixture systems. Further results show that adding process-based simulated biomass as a condition increases the accuracy of the derived phenotypic traits from the predicted images. This demonstrates the potential of our framework to serve as an interface between an image- and process-based crop growth model.
Behind the leaves -- Estimation of occluded grapevine berries with conditional generative adversarial networks
May 21, 2021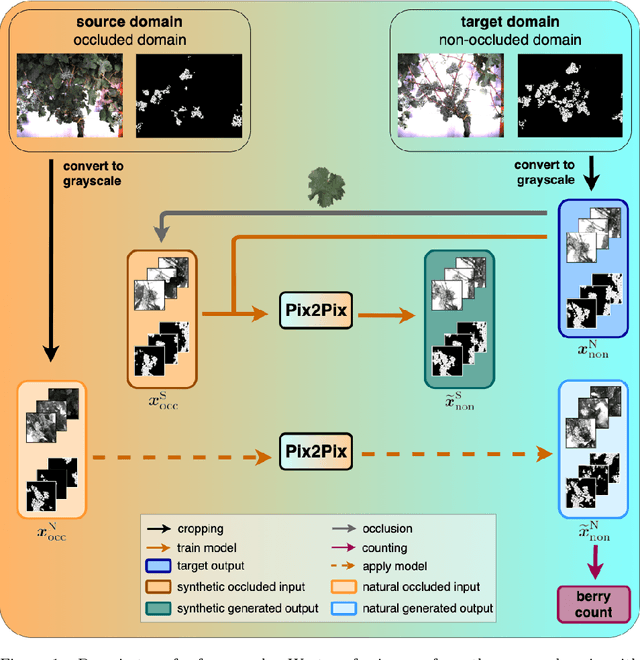
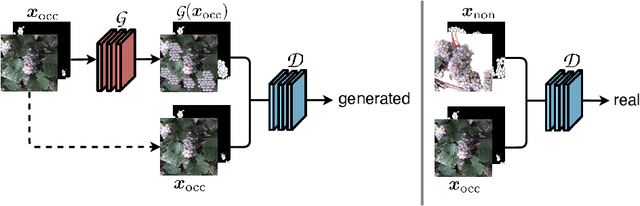
Abstract:The need for accurate yield estimates for viticulture is becoming more important due to increasing competition in the wine market worldwide. One of the most promising methods to estimate the harvest is berry counting, as it can be approached non-destructively, and its process can be automated. In this article, we present a method that addresses the challenge of occluded berries with leaves to obtain a more accurate estimate of the number of berries that will enable a better estimate of the harvest. We use generative adversarial networks, a deep learning-based approach that generates a likely scenario behind the leaves exploiting learned patterns from images with non-occluded berries. Our experiments show that the estimate of the number of berries after applying our method is closer to the manually counted reference. In contrast to applying a factor to the berry count, our approach better adapts to local conditions by directly involving the appearance of the visible berries. Furthermore, we show that our approach can identify which areas in the image should be changed by adding new berries without explicitly requiring information about hidden areas.
Temporal Prediction and Evaluation of Brassica Growth in the Field using Conditional Generative Adversarial Networks
May 17, 2021



Abstract:Farmers frequently assess plant growth and performance as basis for making decisions when to take action in the field, such as fertilization, weed control, or harvesting. The prediction of plant growth is a major challenge, as it is affected by numerous and highly variable environmental factors. This paper proposes a novel monitoring approach that comprises high-throughput imaging sensor measurements and their automatic analysis to predict future plant growth. Our approach's core is a novel machine learning-based growth model based on conditional generative adversarial networks, which is able to predict the future appearance of individual plants. In experiments with RGB time-series images of laboratory-grown Arabidopsis thaliana and field-grown cauliflower plants, we show that our approach produces realistic, reliable, and reasonable images of future growth stages. The automatic interpretation of the generated images through neural network-based instance segmentation allows the derivation of various phenotypic traits that describe plant growth.
Archetypal Analysis for Sparse Representation-based Hyperspectral Sub-pixel Quantification
Feb 08, 2018



Abstract:The estimation of land cover fractions from remote sensing images is a frequently used indicator of the environmental quality. This paper focuses on the quantification of land cover fractions in an urban area of Berlin, Germany, using simulated hyperspectral EnMAP data with a spatial resolution of 30m$\times$30m. We use constrained sparse representation, where each pixel with unknown surface characteristics is expressed by a weighted linear combination of elementary spectra with known land cover class. We automatically determine the elementary spectra from image reference data using archetypal analysis by simplex volume maximization, and combine it with reversible jump Markov chain Monte Carlo method. In our experiments, the estimation of the automatically derived elementary spectra is compared to the estimation obtained by a manually designed spectral library by means of reconstruction error, mean absolute error of the fraction estimates, sum of fractions, $R^2$, and the number of used elementary spectra. The experiments show that a collection of archetypes can be an adequate and efficient alternative to the manually designed spectral library with respect to the mentioned criteria.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge