Jana Kierdorf
Reliability Scores from Saliency Map Clusters for Improved Image-based Harvest-Readiness Prediction in Cauliflower
May 24, 2023

Abstract:Cauliflower is a hand-harvested crop that must fulfill high-quality standards in sales making the timing of harvest important. However, accurately determining harvest-readiness can be challenging due to the cauliflower head being covered by its canopy. While deep learning enables automated harvest-readiness estimation, errors can occur due to field-variability and limited training data. In this paper, we analyze the reliability of a harvest-readiness classifier with interpretable machine learning. By identifying clusters of saliency maps, we derive reliability scores for each classification result using knowledge about the domain and the image properties. For unseen data, the reliability can be used to (i) inform farmers to improve their decision-making and (ii) increase the model prediction accuracy. Using RGB images of single cauliflower plants at different developmental stages from the GrowliFlower dataset, we investigate various saliency mapping approaches and find that they result in different quality of reliability scores. With the most suitable interpretation tool, we adjust the classification result and achieve a 15.72% improvement of the overall accuracy to 88.14% and a 15.44% improvement of the average class accuracy to 88.52% for the GrowliFlower dataset.
GrowliFlower: An image time series dataset for GROWth analysis of cauLIFLOWER
Apr 01, 2022



Abstract:This article presents GrowliFlower, a georeferenced, image-based UAV time series dataset of two monitored cauliflower fields of size 0.39 and 0.60 ha acquired in 2020 and 2021. The dataset contains RGB and multispectral orthophotos from which about 14,000 individual plant coordinates are derived and provided. The coordinates enable the dataset users the extraction of complete and incomplete time series of image patches showing individual plants. The dataset contains collected phenotypic traits of 740 plants, including the developmental stage as well as plant and cauliflower size. As the harvestable product is completely covered by leaves, plant IDs and coordinates are provided to extract image pairs of plants pre and post defoliation, to facilitate estimations of cauliflower head size. Moreover, the dataset contains pixel-accurate leaf and plant instance segmentations, as well as stem annotations to address tasks like classification, detection, segmentation, instance segmentation, and similar computer vision tasks. The dataset aims to foster the development and evaluation of machine learning approaches. It specifically focuses on the analysis of growth and development of cauliflower and the derivation of phenotypic traits to foster the development of automation in agriculture. Two baseline results of instance segmentation at plant and leaf level based on the labeled instance segmentation data are presented. The entire data set is publicly available.
Agricultural Plant Cataloging and Establishment of a Data Framework from UAV-based Crop Images by Computer Vision
Jan 11, 2022



Abstract:UAV-based image retrieval in modern agriculture enables gathering large amounts of spatially referenced crop image data. In large-scale experiments, however, UAV images suffer from containing a multitudinous amount of crops in a complex canopy architecture. Especially for the observation of temporal effects, this complicates the recognition of individual plants over several images and the extraction of relevant information tremendously. In this work, we present a hands-on workflow for the automatized temporal and spatial identification and individualization of crop images from UAVs abbreviated as "cataloging" based on comprehensible computer vision methods. We evaluate the workflow on two real-world datasets. One dataset is recorded for observation of Cercospora leaf spot - a fungal disease - in sugar beet over an entire growing cycle. The other one deals with harvest prediction of cauliflower plants. The plant catalog is utilized for the extraction of single plant images seen over multiple time points. This gathers large-scale spatio-temporal image dataset that in turn can be applied to train further machine learning models including various data layers. The presented approach improves analysis and interpretation of UAV data in agriculture significantly. By validation with some reference data, our method shows an accuracy that is similar to more complex deep learning-based recognition techniques. Our workflow is able to automatize plant cataloging and training image extraction, especially for large datasets.
Behind the leaves -- Estimation of occluded grapevine berries with conditional generative adversarial networks
May 21, 2021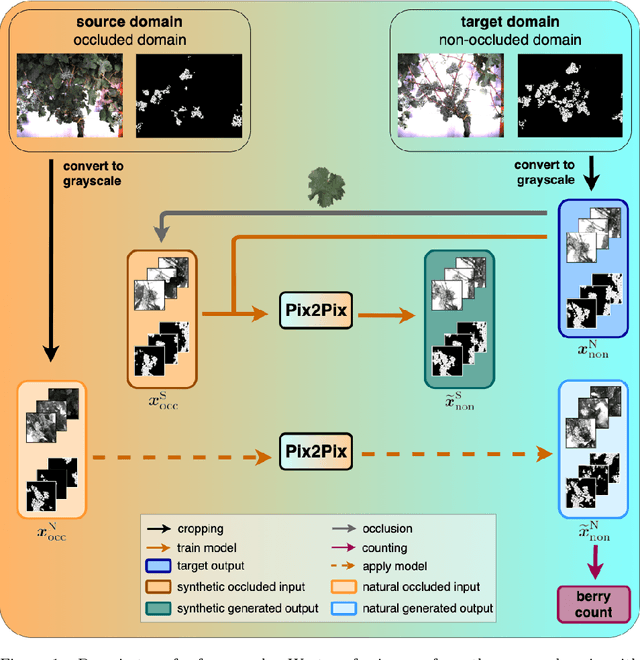
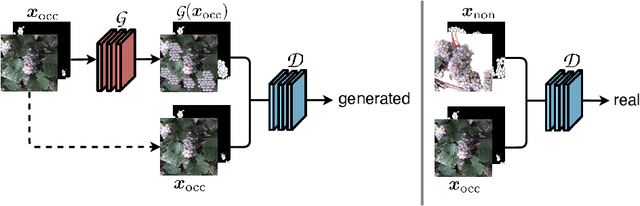
Abstract:The need for accurate yield estimates for viticulture is becoming more important due to increasing competition in the wine market worldwide. One of the most promising methods to estimate the harvest is berry counting, as it can be approached non-destructively, and its process can be automated. In this article, we present a method that addresses the challenge of occluded berries with leaves to obtain a more accurate estimate of the number of berries that will enable a better estimate of the harvest. We use generative adversarial networks, a deep learning-based approach that generates a likely scenario behind the leaves exploiting learned patterns from images with non-occluded berries. Our experiments show that the estimate of the number of berries after applying our method is closer to the manually counted reference. In contrast to applying a factor to the berry count, our approach better adapts to local conditions by directly involving the appearance of the visible berries. Furthermore, we show that our approach can identify which areas in the image should be changed by adding new berries without explicitly requiring information about hidden areas.
Temporal Prediction and Evaluation of Brassica Growth in the Field using Conditional Generative Adversarial Networks
May 17, 2021



Abstract:Farmers frequently assess plant growth and performance as basis for making decisions when to take action in the field, such as fertilization, weed control, or harvesting. The prediction of plant growth is a major challenge, as it is affected by numerous and highly variable environmental factors. This paper proposes a novel monitoring approach that comprises high-throughput imaging sensor measurements and their automatic analysis to predict future plant growth. Our approach's core is a novel machine learning-based growth model based on conditional generative adversarial networks, which is able to predict the future appearance of individual plants. In experiments with RGB time-series images of laboratory-grown Arabidopsis thaliana and field-grown cauliflower plants, we show that our approach produces realistic, reliable, and reasonable images of future growth stages. The automatic interpretation of the generated images through neural network-based instance segmentation allows the derivation of various phenotypic traits that describe plant growth.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge