Luis Carlos Rivera Monroy
Comparative Analysis of Radiomic Features and Gene Expression Profiles in Histopathology Data Using Graph Neural Networks
Dec 25, 2023Abstract:This study leverages graph neural networks to integrate MELC data with Radiomic-extracted features for melanoma classification, focusing on cell-wise analysis. It assesses the effectiveness of gene expression profiles and Radiomic features, revealing that Radiomic features, particularly when combined with UMAP for dimensionality reduction, significantly enhance classification performance. Notably, using Radiomics contributes to increased diagnostic accuracy and computational efficiency, as it allows for the extraction of critical data from fewer stains, thereby reducing operational costs. This methodology marks an advancement in computational dermatology for melanoma cell classification, setting the stage for future research and potential developments.
Employing Graph Representations for Cell-level Characterization of Melanoma MELC Samples
Nov 10, 2022


Abstract:Histopathology imaging is crucial for the diagnosis and treatment of skin diseases. For this reason, computer-assisted approaches have gained popularity and shown promising results in tasks such as segmentation and classification of skin disorders. However, collecting essential data and sufficiently high-quality annotations is a challenge. This work describes a pipeline that uses suspected melanoma samples that have been characterized using Multi-Epitope-Ligand Cartography (MELC). This cellular-level tissue characterisation is then represented as a graph and used to train a graph neural network. This imaging technology, combined with the methodology proposed in this work, achieves a classification accuracy of 87%, outperforming existing approaches by 10%.
Multi-Channel Volumetric Neural Network for Knee Cartilage Segmentation in Cone-beam CT
Dec 03, 2019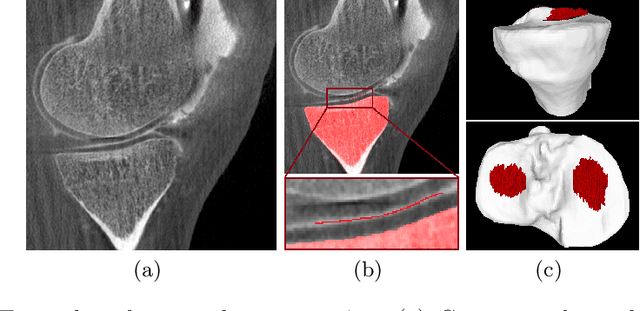
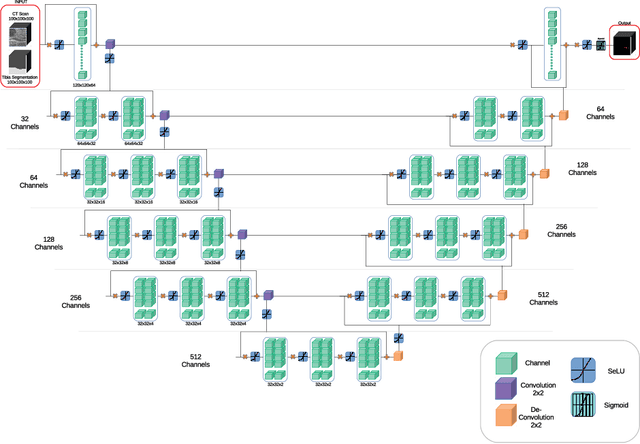
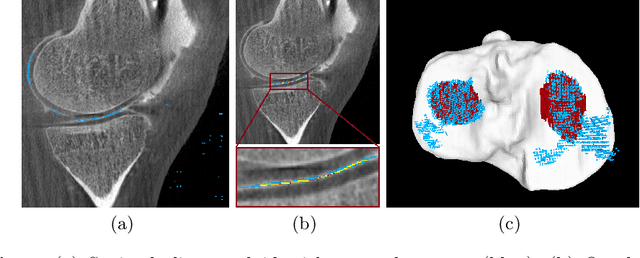
Abstract:Analyzing knee cartilage thickness and strain under load can help to further the understanding of the effects of diseases like Osteoarthritis. A precise segmentation of the cartilage is a necessary prerequisite for this analysis. This segmentation task has mainly been addressed in Magnetic Resonance Imaging, and was rarely investigated on contrast-enhanced Computed Tomography, where contrast agent visualizes the border between femoral and tibial cartilage. To overcome the main drawback of manual segmentation, namely its high time investment, we propose to use a 3D Convolutional Neural Network for this task. The presented architecture consists of a V-Net with SeLu activation, and a Tversky loss function. Due to the high imbalance between very few cartilage pixels and many background pixels, a high false positive rate is to be expected. To reduce this rate, the two largest segmented point clouds are extracted using a connected component analysis, since they most likely represent the medial and lateral tibial cartilage surfaces. The resulting segmentations are compared to manual segmentations, and achieve on average a recall of 0.69, which confirms the feasibility of this approach.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge