Lufan Chang
Lightweight Method for Interactive 3D Medical Image Segmentation with Multi-Round Result Fusion
Dec 11, 2024Abstract:In medical imaging, precise annotation of lesions or organs is often required. However, 3D volumetric images typically consist of hundreds or thousands of slices, making the annotation process extremely time-consuming and laborious. Recently, the Segment Anything Model (SAM) has drawn widespread attention due to its remarkable zero-shot generalization capabilities in interactive segmentation. While researchers have explored adapting SAM for medical applications, such as using SAM adapters or constructing 3D SAM models, a key question remains: Can traditional CNN networks achieve the same strong zero-shot generalization in this task? In this paper, we propose the Lightweight Interactive Network for 3D Medical Image Segmentation (LIM-Net), a novel approach demonstrating the potential of compact CNN-based models. Built upon a 2D CNN backbone, LIM-Net initiates segmentation by generating a 2D prompt mask from user hints. This mask is then propagated through the 3D sequence via the Memory Module. To refine and stabilize results during interaction, the Multi-Round Result Fusion (MRF) Module selects and merges optimal masks from multiple rounds. Our extensive experiments across multiple datasets and modalities demonstrate LIM-Net's competitive performance. It exhibits stronger generalization to unseen data compared to SAM-based models, with competitive accuracy while requiring fewer interactions. Notably, LIM-Net's lightweight design offers significant advantages in deployment and inference efficiency, with low GPU memory consumption suitable for resource-constrained environments. These promising results demonstrate LIM-Net can serve as a strong baseline, complementing and contrasting with popular SAM models to further boost effective interactive medical image segmentation. The code will be released at \url{https://github.com/goodtime-123/LIM-Net}.
DARWIN: A Highly Flexible Platform for Imaging Research in Radiology
Sep 02, 2020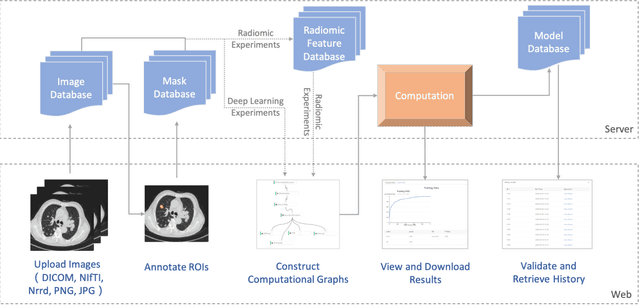
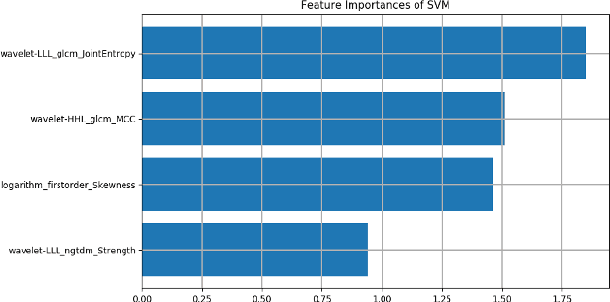
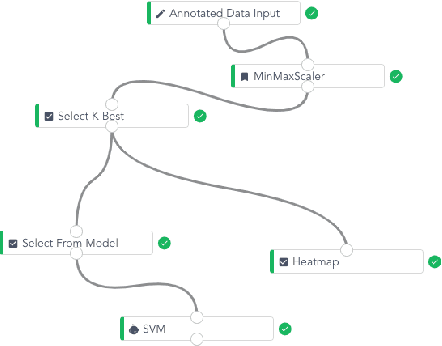
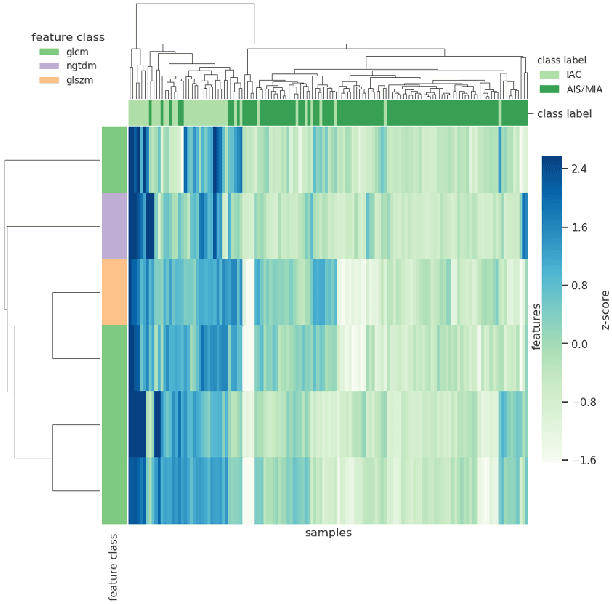
Abstract:To conduct a radiomics or deep learning research experiment, the radiologists or physicians need to grasp the needed programming skills, which, however, could be frustrating and costly when they have limited coding experience. In this paper, we present DARWIN, a flexible research platform with a graphical user interface for medical imaging research. Our platform is consists of a radiomics module and a deep learning module. The radiomics module can extract more than 1000 dimension features(first-, second-, and higher-order) and provided many draggable supervised and unsupervised machine learning models. Our deep learning module integrates state of the art architectures of classification, detection, and segmentation tasks. It allows users to manually select hyperparameters, or choose an algorithm to automatically search for the best ones. DARWIN also offers the possibility for users to define a custom pipeline for their experiment. These flexibilities enable radiologists to carry out various experiments easily.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge