Lou Scheffer
A Context-aware Delayed Agglomeration Framework for Electron Microscopy Segmentation
Mar 23, 2015



Abstract:Electron Microscopy (EM) image (or volume) segmentation has become significantly important in recent years as an instrument for connectomics. This paper proposes a novel agglomerative framework for EM segmentation. In particular, given an over-segmented image or volume, we propose a novel framework for accurately clustering regions of the same neuron. Unlike existing agglomerative methods, the proposed context-aware algorithm divides superpixels (over-segmented regions) of different biological entities into different subsets and agglomerates them separately. In addition, this paper describes a "delayed" scheme for agglomerative clustering that postpones some of the merge decisions, pertaining to newly formed bodies, in order to generate a more confident boundary prediction. We report significant improvements attained by the proposed approach in segmentation accuracy over existing standard methods on 2D and 3D datasets.
Super-resolution using Sparse Representations over Learned Dictionaries: Reconstruction of Brain Structure using Electron Microscopy
Oct 01, 2012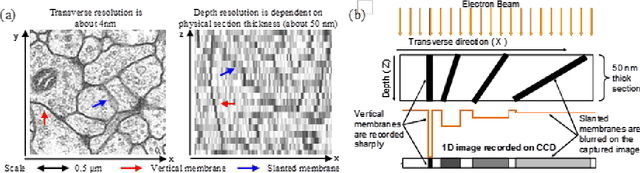
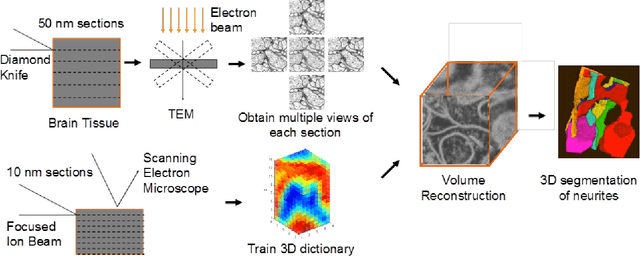
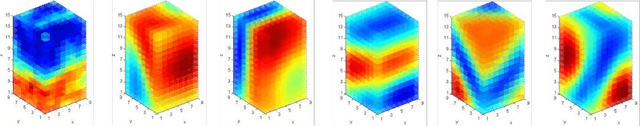
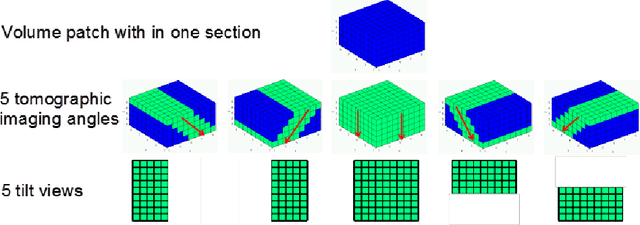
Abstract:A central problem in neuroscience is reconstructing neuronal circuits on the synapse level. Due to a wide range of scales in brain architecture such reconstruction requires imaging that is both high-resolution and high-throughput. Existing electron microscopy (EM) techniques possess required resolution in the lateral plane and either high-throughput or high depth resolution but not both. Here, we exploit recent advances in unsupervised learning and signal processing to obtain high depth-resolution EM images computationally without sacrificing throughput. First, we show that the brain tissue can be represented as a sparse linear combination of localized basis functions that are learned using high-resolution datasets. We then develop compressive sensing-inspired techniques that can reconstruct the brain tissue from very few (typically 5) tomographic views of each section. This enables tracing of neuronal processes and, hence, high throughput reconstruction of neural circuits on the level of individual synapses.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge