Longhua Qian
Multi-layer Sequence Labeling-based Joint Biomedical Event Extraction
Aug 14, 2024


Abstract:In recent years, biomedical event extraction has been dominated by complicated pipeline and joint methods, which need to be simplified. In addition, existing work has not effectively utilized trigger word information explicitly. Hence, we propose MLSL, a method based on multi-layer sequence labeling for joint biomedical event extraction. MLSL does not introduce prior knowledge and complex structures. Moreover, it explicitly incorporates the information of candidate trigger words into the sequence labeling to learn the interaction relationships between trigger words and argument roles. Based on this, MLSL can learn well with just a simple workflow. Extensive experimentation demonstrates the superiority of MLSL in terms of extraction performance compared to other state-of-the-art methods.
Pipelined Biomedical Event Extraction Rivaling Joint Learning
Mar 19, 2024


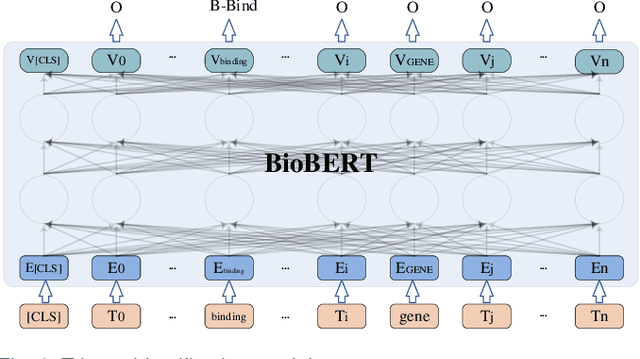
Abstract:Biomedical event extraction is an information extraction task to obtain events from biomedical text, whose targets include the type, the trigger, and the respective arguments involved in an event. Traditional biomedical event extraction usually adopts a pipelined approach, which contains trigger identification, argument role recognition, and finally event construction either using specific rules or by machine learning. In this paper, we propose an n-ary relation extraction method based on the BERT pre-training model to construct Binding events, in order to capture the semantic information about an event's context and its participants. The experimental results show that our method achieves promising results on the GE11 and GE13 corpora of the BioNLP shared task with F1 scores of 63.14% and 59.40%, respectively. It demonstrates that by significantly improving theperformance of Binding events, the overall performance of the pipelined event extraction approach or even exceeds those of current joint learning methods.
Joint Learning-based Causal Relation Extraction from Biomedical Literature
Aug 02, 2022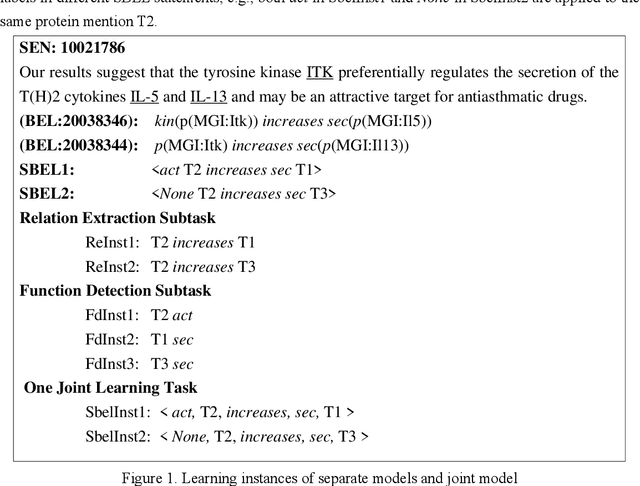
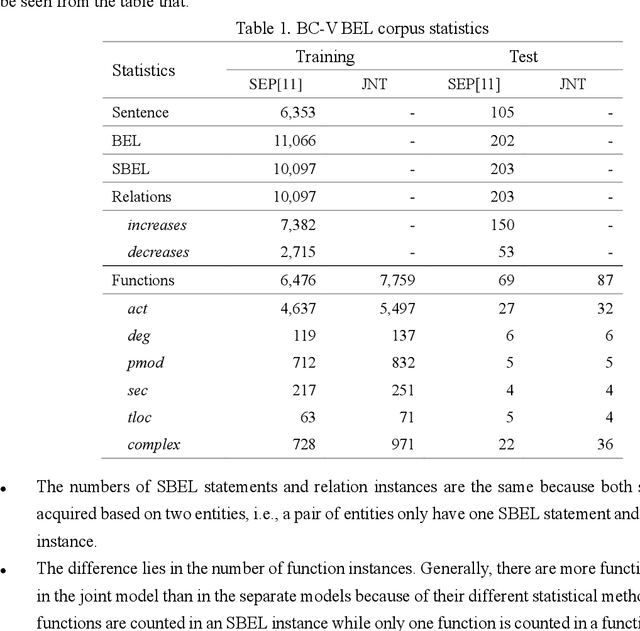
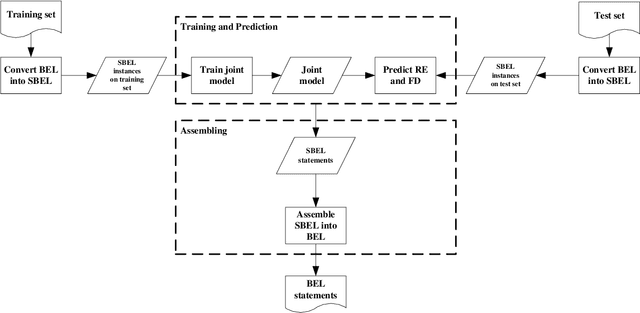
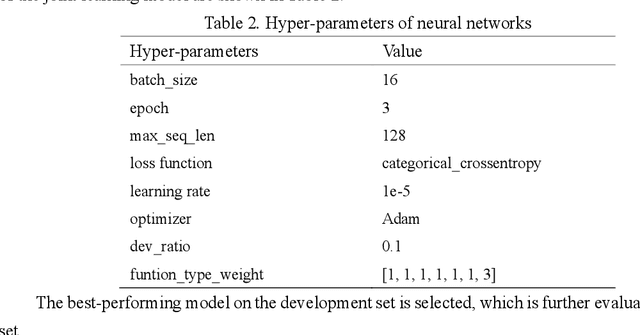
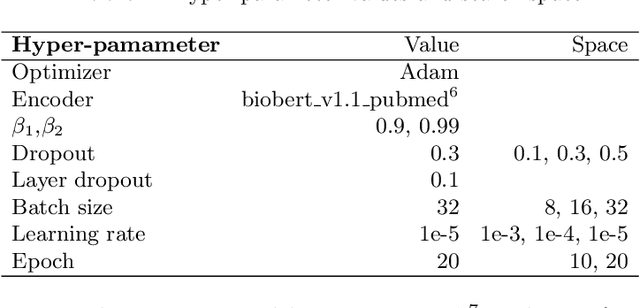
Abstract:Causal relation extraction of biomedical entities is one of the most complex tasks in biomedical text mining, which involves two kinds of information: entity relations and entity functions. One feasible approach is to take relation extraction and function detection as two independent sub-tasks. However, this separate learning method ignores the intrinsic correlation between them and leads to unsatisfactory performance. In this paper, we propose a joint learning model, which combines entity relation extraction and entity function detection to exploit their commonality and capture their inter-relationship, so as to improve the performance of biomedical causal relation extraction. Meanwhile, during the model training stage, different function types in the loss function are assigned different weights. Specifically, the penalty coefficient for negative function instances increases to effectively improve the precision of function detection. Experimental results on the BioCreative-V Track 4 corpus show that our joint learning model outperforms the separate models in BEL statement extraction, achieving the F1 scores of 58.4% and 37.3% on the test set in Stage 2 and Stage 1 evaluations, respectively. This demonstrates that our joint learning system reaches the state-of-the-art performance in Stage 2 compared with other systems.
Modeling Graph Structure in Transformer for Better AMR-to-Text Generation
Aug 31, 2019



Abstract:Recent studies on AMR-to-text generation often formalize the task as a sequence-to-sequence (seq2seq) learning problem by converting an Abstract Meaning Representation (AMR) graph into a word sequence. Graph structures are further modeled into the seq2seq framework in order to utilize the structural information in the AMR graphs. However, previous approaches only consider the relations between directly connected concepts while ignoring the rich structure in AMR graphs. In this paper we eliminate such a strong limitation and propose a novel structure-aware self-attention approach to better modeling the relations between indirectly connected concepts in the state-of-the-art seq2seq model, i.e., the Transformer. In particular, a few different methods are explored to learn structural representations between two concepts. Experimental results on English AMR benchmark datasets show that our approach significantly outperforms the state of the art with 29.66 and 31.82 BLEU scores on LDC2015E86 and LDC2017T10, respectively. To the best of our knowledge, these are the best results achieved so far by supervised models on the benchmarks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge