Lituan Wang
Optical Flow Representation Alignment Mamba Diffusion Model for Medical Video Generation
Nov 03, 2024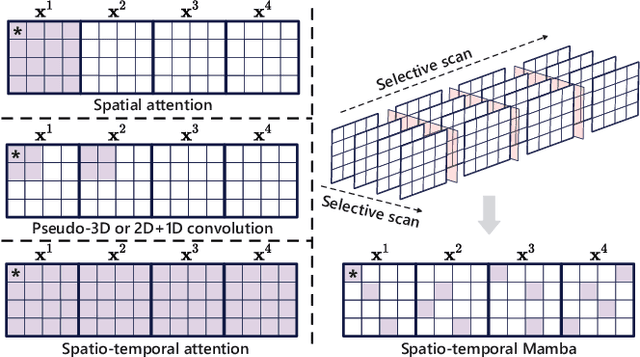
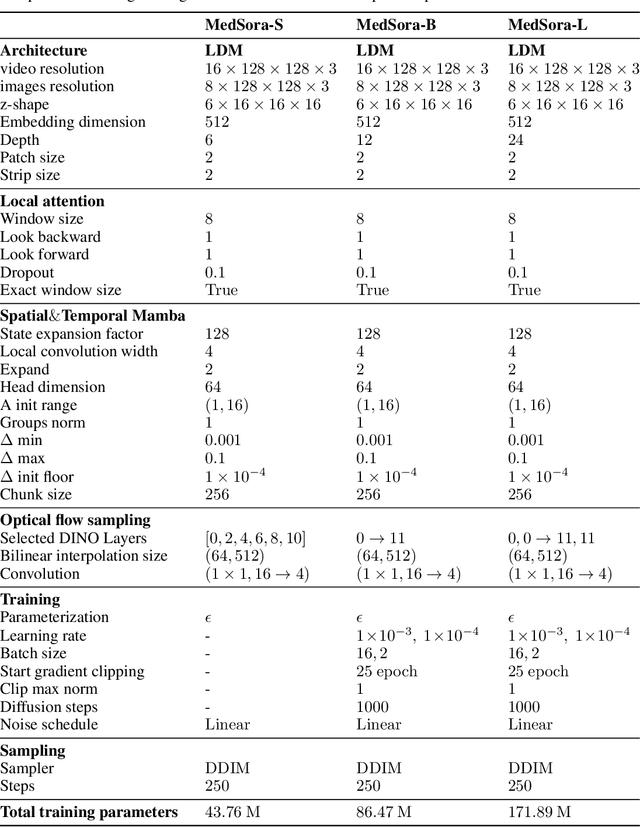
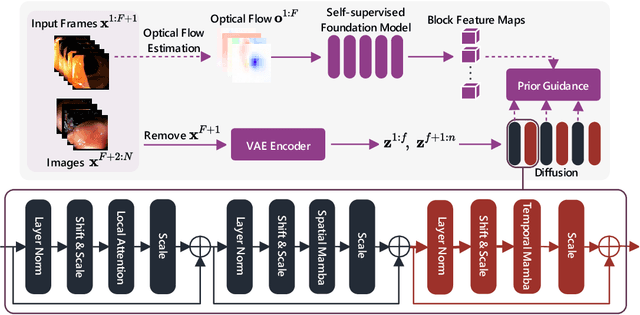
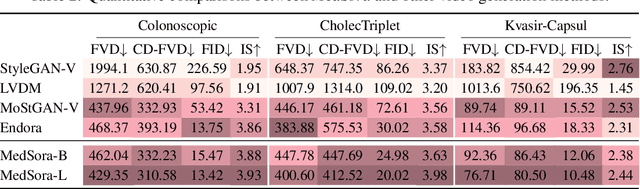
Abstract:Medical video generation models are expected to have a profound impact on the healthcare industry, including but not limited to medical education and training, surgical planning, and simulation. Current video diffusion models typically build on image diffusion architecture by incorporating temporal operations (such as 3D convolution and temporal attention). Although this approach is effective, its oversimplification limits spatio-temporal performance and consumes substantial computational resources. To counter this, we propose Medical Simulation Video Generator (MedSora), which incorporates three key elements: i) a video diffusion framework integrates the advantages of attention and Mamba, balancing low computational load with high-quality video generation, ii) an optical flow representation alignment method that implicitly enhances attention to inter-frame pixels, and iii) a video variational autoencoder (VAE) with frequency compensation addresses the information loss of medical features that occurs when transforming pixel space into latent features and then back to pixel frames. Extensive experiments and applications demonstrate that MedSora exhibits superior visual quality in generating medical videos, outperforming the most advanced baseline methods. Further results and code are available at https://wongzbb.github.io/MedSora
Soft Masked Mamba Diffusion Model for CT to MRI Conversion
Jun 22, 2024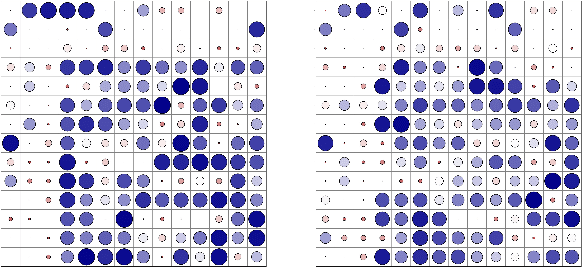
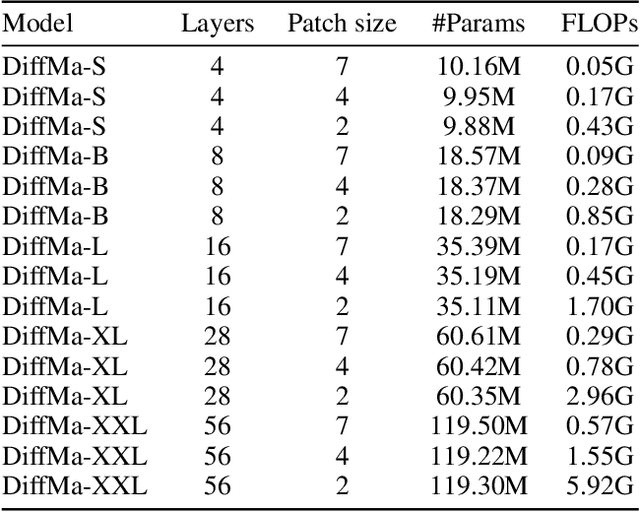
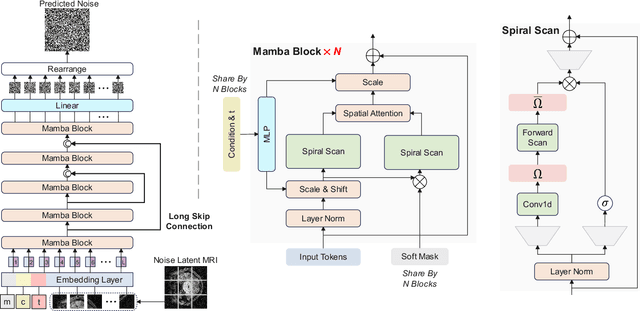
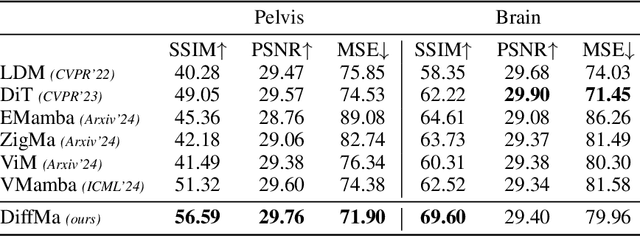
Abstract:Magnetic Resonance Imaging (MRI) and Computed Tomography (CT) are the predominant modalities utilized in the field of medical imaging. Although MRI capture the complexity of anatomical structures with greater detail than CT, it entails a higher financial costs and requires longer image acquisition times. In this study, we aim to train latent diffusion model for CT to MRI conversion, replacing the commonly-used U-Net or Transformer backbone with a State-Space Model (SSM) called Mamba that operates on latent patches. First, we noted critical oversights in the scan scheme of most Mamba-based vision methods, including inadequate attention to the spatial continuity of patch tokens and the lack of consideration for their varying importance to the target task. Secondly, extending from this insight, we introduce Diffusion Mamba (DiffMa), employing soft masked to integrate Cross-Sequence Attention into Mamba and conducting selective scan in a spiral manner. Lastly, extensive experiments demonstrate impressive performance by DiffMa in medical image generation tasks, with notable advantages in input scaling efficiency over existing benchmark models. The code and models are available at https://github.com/wongzbb/DiffMa-Diffusion-Mamba
PCLMix: Weakly Supervised Medical Image Segmentation via Pixel-Level Contrastive Learning and Dynamic Mix Augmentation
May 10, 2024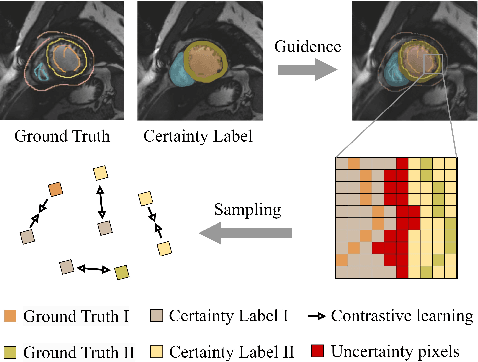
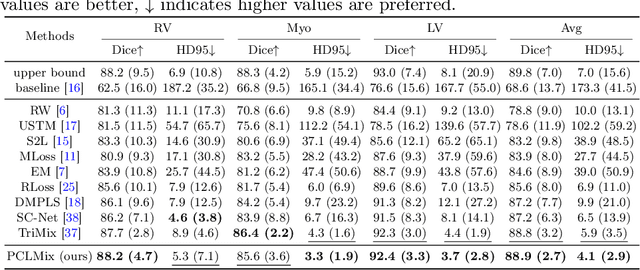
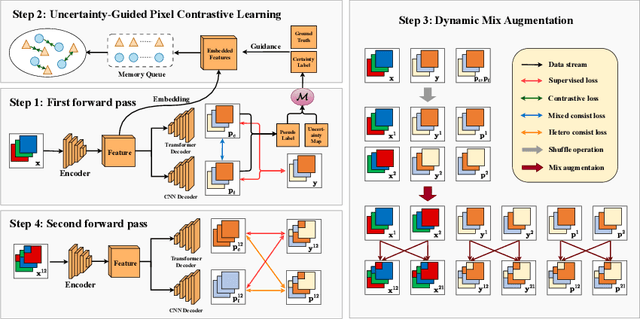
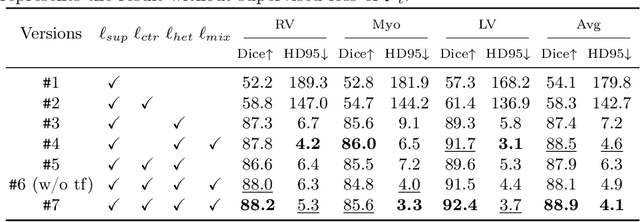
Abstract:In weakly supervised medical image segmentation, the absence of structural priors and the discreteness of class feature distribution present a challenge, i.e., how to accurately propagate supervision signals from local to global regions without excessively spreading them to other irrelevant regions? To address this, we propose a novel weakly supervised medical image segmentation framework named PCLMix, comprising dynamic mix augmentation, pixel-level contrastive learning, and consistency regularization strategies. Specifically, PCLMix is built upon a heterogeneous dual-decoder backbone, addressing the absence of structural priors through a strategy of dynamic mix augmentation during training. To handle the discrete distribution of class features, PCLMix incorporates pixel-level contrastive learning based on prediction uncertainty, effectively enhancing the model's ability to differentiate inter-class pixel differences and intra-class consistency. Furthermore, to reinforce segmentation consistency and robustness, PCLMix employs an auxiliary decoder for dual consistency regularization. In the inference phase, the auxiliary decoder will be dropped and no computation complexity is increased. Extensive experiments on the ACDC dataset demonstrate that PCLMix appropriately propagates local supervision signals to the global scale, further narrowing the gap between weakly supervised and fully supervised segmentation methods. Our code is available at https://github.com/Torpedo2648/PCLMix.
LanDA: Language-Guided Multi-Source Domain Adaptation
Jan 25, 2024Abstract:Multi-Source Domain Adaptation (MSDA) aims to mitigate changes in data distribution when transferring knowledge from multiple labeled source domains to an unlabeled target domain. However, existing MSDA techniques assume target domain images are available, yet overlook image-rich semantic information. Consequently, an open question is whether MSDA can be guided solely by textual cues in the absence of target domain images. By employing a multimodal model with a joint image and language embedding space, we propose a novel language-guided MSDA approach, termed LanDA, based on optimal transfer theory, which facilitates the transfer of multiple source domains to a new target domain, requiring only a textual description of the target domain without needing even a single target domain image, while retaining task-relevant information. We present extensive experiments across different transfer scenarios using a suite of relevant benchmarks, demonstrating that LanDA outperforms standard fine-tuning and ensemble approaches in both target and source domains.
Auto Machine Learning for Medical Image Analysis by Unifying the Search on Data Augmentation and Neural Architecture
Jul 21, 2022



Abstract:Automated data augmentation, which aims at engineering augmentation policy automatically, recently draw a growing research interest. Many previous auto-augmentation methods utilized a Density Matching strategy by evaluating policies in terms of the test-time augmentation performance. In this paper, we theoretically and empirically demonstrated the inconsistency between the train and validation set of small-scale medical image datasets, referred to as in-domain sampling bias. Next, we demonstrated that the in-domain sampling bias might cause the inefficiency of Density Matching. To address the problem, an improved augmentation search strategy, named Augmented Density Matching, was proposed by randomly sampling policies from a prior distribution for training. Moreover, an efficient automatical machine learning(AutoML) algorithm was proposed by unifying the search on data augmentation and neural architecture. Experimental results indicated that the proposed methods outperformed state-of-the-art approaches on MedMNIST, a pioneering benchmark designed for AutoML in medical image analysis.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge