Laurence Yang
GripNet: Graph Information Propagation on Supergraph for Heterogeneous Graphs
Oct 29, 2020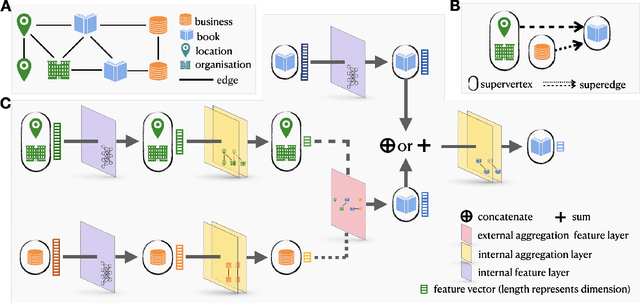



Abstract:Heterogeneous graph representation learning aims to learn low-dimensional vector representations of different types of entities and relations to empower downstream tasks. Existing methods either capture semantic relationships but indirectly leverage node/edge attributes in a complex way, or leverage node/edge attributes directly without taking semantic relationships into account. When involving multiple convolution operations, they also have poor scalability. To overcome these limitations, this paper proposes a flexible and efficient Graph information propagation Network (GripNet) framework. Specifically, we introduce a new supergraph data structure consisting of supervertices and superedges. A supervertex is a semantically-coherent subgraph. A superedge defines an information propagation path between two supervertices. GripNet learns new representations for the supervertex of interest by propagating information along the defined path using multiple layers. We construct multiple large-scale graphs and evaluate GripNet against competing methods to show its superiority in link prediction, node classification, and data integration.
Genome-scale estimation of cellular objectives
Oct 15, 2018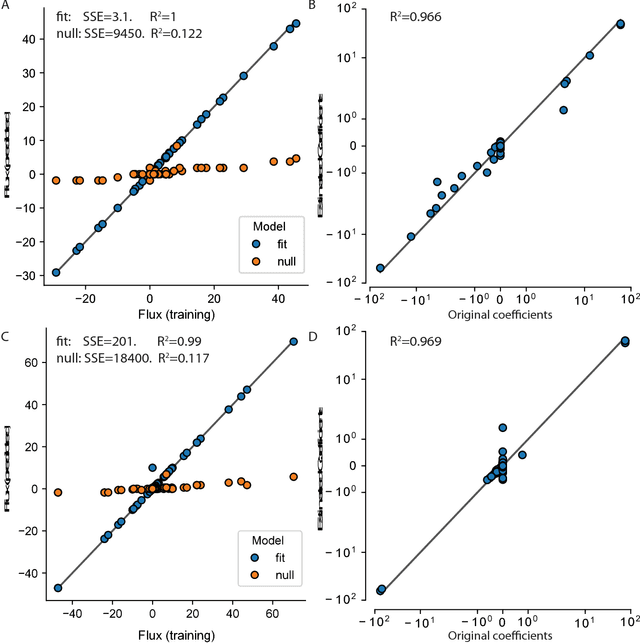
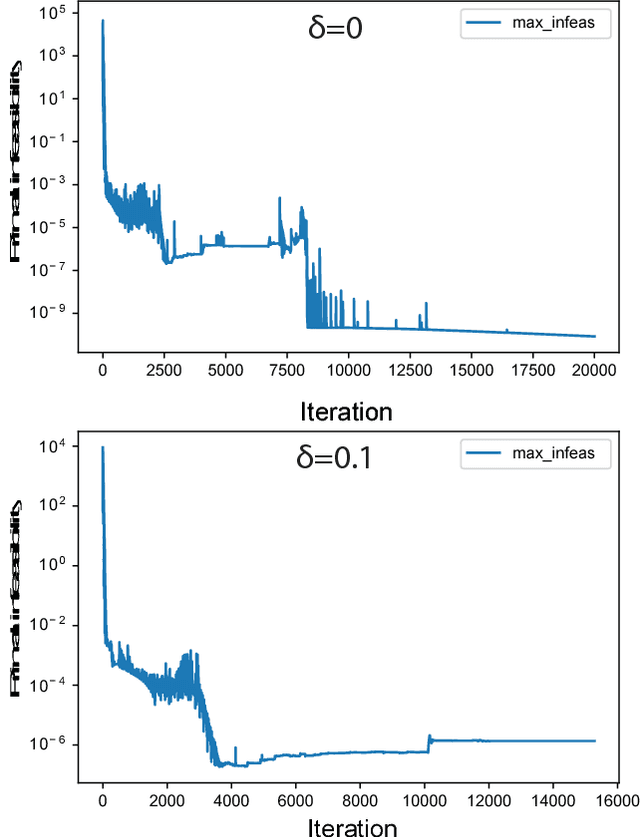
Abstract:Cellular metabolism is predicted accurately at the genome-scale using constraint based modeling. Such predictions typically rely on optimizing an assumed cellular objective function, which takes the form of a stoichiometrically-determined reaction such as biomass synthesis, ATP yield, or reactive oxygen species formation. While these objective functions are typically constructed by hand, several algorithms have been developed to estimate them from data. Generally, two approaches for data-driven objective estimation exist: estimating objective weights for existing reactions, and de novo generation of a new objective reaction. The latter approach can discover objectives that are not describable as a linear combination of existing reactions. However, it requires solving a nonconvex optimization problem and its scalability to genome-scale models has not been demonstrated. Here, we develop a new algorithm that extends existing approaches for de novo objective generation and solve it using the alternating direction method of multipliers (ADMM). We demonstrate our approach on a genome-scale model and show that it identifies de novo objectives from measured fluxes with tunable sparsity.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge