Genome-scale estimation of cellular objectives
Paper and Code
Oct 15, 2018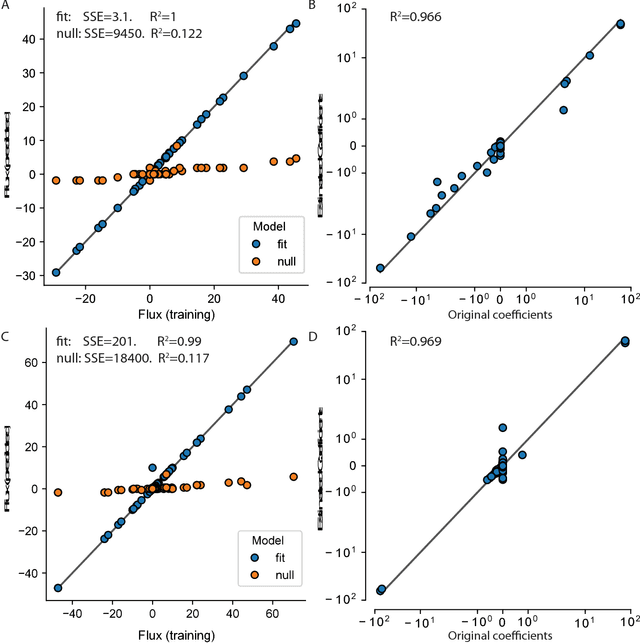
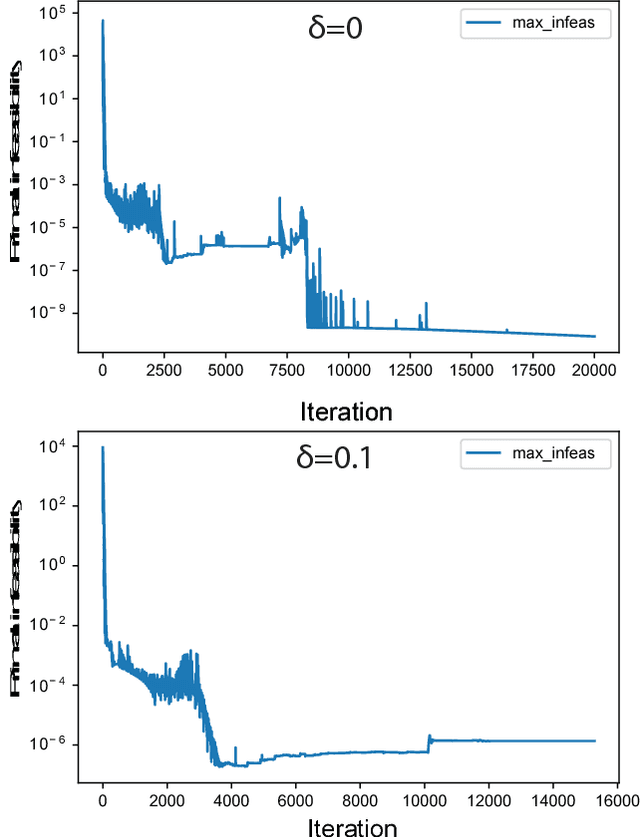
Cellular metabolism is predicted accurately at the genome-scale using constraint based modeling. Such predictions typically rely on optimizing an assumed cellular objective function, which takes the form of a stoichiometrically-determined reaction such as biomass synthesis, ATP yield, or reactive oxygen species formation. While these objective functions are typically constructed by hand, several algorithms have been developed to estimate them from data. Generally, two approaches for data-driven objective estimation exist: estimating objective weights for existing reactions, and de novo generation of a new objective reaction. The latter approach can discover objectives that are not describable as a linear combination of existing reactions. However, it requires solving a nonconvex optimization problem and its scalability to genome-scale models has not been demonstrated. Here, we develop a new algorithm that extends existing approaches for de novo objective generation and solve it using the alternating direction method of multipliers (ADMM). We demonstrate our approach on a genome-scale model and show that it identifies de novo objectives from measured fluxes with tunable sparsity.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge