Shengqi Sang
GripNet: Graph Information Propagation on Supergraph for Heterogeneous Graphs
Oct 29, 2020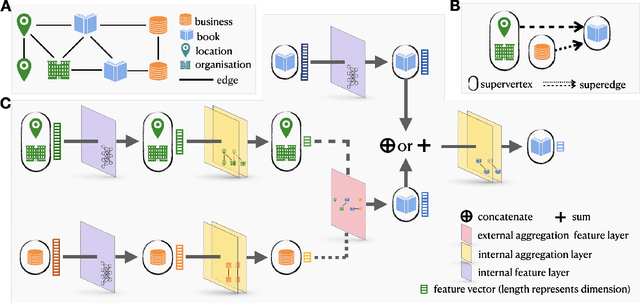



Abstract:Heterogeneous graph representation learning aims to learn low-dimensional vector representations of different types of entities and relations to empower downstream tasks. Existing methods either capture semantic relationships but indirectly leverage node/edge attributes in a complex way, or leverage node/edge attributes directly without taking semantic relationships into account. When involving multiple convolution operations, they also have poor scalability. To overcome these limitations, this paper proposes a flexible and efficient Graph information propagation Network (GripNet) framework. Specifically, we introduce a new supergraph data structure consisting of supervertices and superedges. A supervertex is a semantically-coherent subgraph. A superedge defines an information propagation path between two supervertices. GripNet learns new representations for the supervertex of interest by propagating information along the defined path using multiple layers. We construct multiple large-scale graphs and evaluate GripNet against competing methods to show its superiority in link prediction, node classification, and data integration.
Tri-graph Information Propagation for Polypharmacy Side Effect Prediction
Jan 28, 2020



Abstract:The use of drug combinations often leads to polypharmacy side effects (POSE). A recent method formulates POSE prediction as a link prediction problem on a graph of drugs and proteins, and solves it with Graph Convolutional Networks (GCNs). However, due to the complex relationships in POSE, this method has high computational cost and memory demand. This paper proposes a flexible Tri-graph Information Propagation (TIP) model that operates on three subgraphs to learn representations progressively by propagation from protein-protein graph to drug-drug graph via protein-drug graph. Experiments show that TIP improves accuracy by 7%+, time efficiency by 83$\times$, and space efficiency by 3$\times$.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge