Kristy K. Brock
The University of Texas MD Anderson Cancer Center
Importance of Feature Extraction in the Calculation of Fréchet Distance for Medical Imaging
Nov 22, 2023Abstract:Fr\'echet Inception Distance is a widely used metric for evaluating synthetic image quality that utilizes an ImageNet-trained InceptionV3 network as a feature extractor. However, its application in medical imaging lacks a standard feature extractor, leading to biased and inconsistent comparisons. This study aimed to compare state-of-the-art feature extractors for computing Fr\'echet Distances (FDs) in medical imaging. A StyleGAN2 network was trained with data augmentation techniques tailored for limited data domains on datasets comprising three medical imaging modalities and four anatomical locations. Human evaluation of generative quality (via a visual Turing test) was compared to FDs calculated using ImageNet-trained InceptionV3, ResNet50, SwAV, DINO, and Swin Transformer architectures, in addition to an InceptionV3 network trained on a large medical dataset, RadImageNet. All ImageNet-based extractors were consistent with each other, but only SwAV was significantly correlated with medical expert judgment. The RadImageNet-based FD showed volatility and lacked correlation with human judgment. Caution is advised when using medical image-trained extraction networks in the FD calculation. These networks should be rigorously evaluated on the imaging modality under consideration and publicly released. ImageNet-based extractors, while imperfect, are consistent and widely understood. Training extraction networks with SwAV is a promising approach for synthetic medical image evaluation.
Dimensionality Reduction for Improving Out-of-Distribution Detection in Medical Image Segmentation
Aug 07, 2023Abstract:Clinically deployed segmentation models are known to fail on data outside of their training distribution. As these models perform well on most cases, it is imperative to detect out-of-distribution (OOD) images at inference to protect against automation bias. This work applies the Mahalanobis distance post hoc to the bottleneck features of a Swin UNETR model that segments the liver on T1-weighted magnetic resonance imaging. By reducing the dimensions of the bottleneck features with principal component analysis, OOD images were detected with high performance and minimal computational load.
StyleGAN2-based Out-of-Distribution Detection for Medical Imaging
Jul 10, 2023
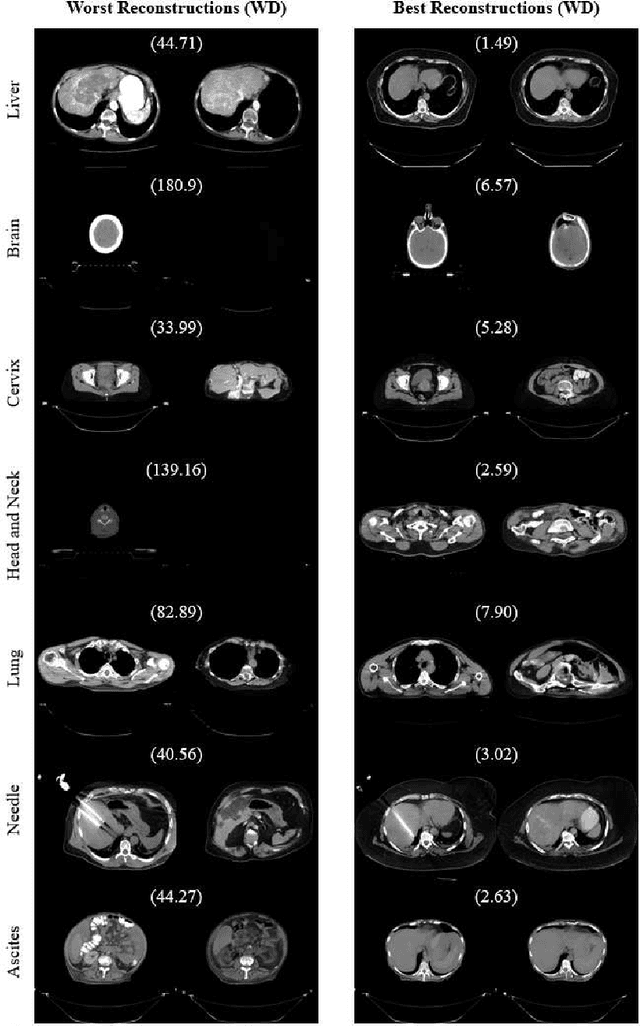
Abstract:One barrier to the clinical deployment of deep learning-based models is the presence of images at runtime that lie far outside the training distribution of a given model. We aim to detect these out-of-distribution (OOD) images with a generative adversarial network (GAN). Our training dataset was comprised of 3,234 liver-containing computed tomography (CT) scans from 456 patients. Our OOD test data consisted of CT images of the brain, head and neck, lung, cervix, and abnormal livers. A StyleGAN2-ADA architecture was employed to model the training distribution. Images were reconstructed using backpropagation. Reconstructions were evaluated using the Wasserstein distance, mean squared error, and the structural similarity index measure. OOD detection was evaluated with the area under the receiver operating characteristic curve (AUROC). Our paradigm distinguished between liver and non-liver CT with greater than 90% AUROC. It was also completely unable to reconstruct liver artifacts, such as needles and ascites.
* Extended abstract published in the "Medical Imaging Meets NeurIPS" workshop at NeurIPS 2022. Original abstract can be found at http://www.cse.cuhk.edu.hk/~qdou/public/medneurips2022/125.pdf
Evaluating the Performance of StyleGAN2-ADA on Medical Images
Oct 07, 2022Abstract:Although generative adversarial networks (GANs) have shown promise in medical imaging, they have four main limitations that impeded their utility: computational cost, data requirements, reliable evaluation measures, and training complexity. Our work investigates each of these obstacles in a novel application of StyleGAN2-ADA to high-resolution medical imaging datasets. Our dataset is comprised of liver-containing axial slices from non-contrast and contrast-enhanced computed tomography (CT) scans. Additionally, we utilized four public datasets composed of various imaging modalities. We trained a StyleGAN2 network with transfer learning (from the Flickr-Faces-HQ dataset) and data augmentation (horizontal flipping and adaptive discriminator augmentation). The network's generative quality was measured quantitatively with the Fr\'echet Inception Distance (FID) and qualitatively with a visual Turing test given to seven radiologists and radiation oncologists. The StyleGAN2-ADA network achieved a FID of 5.22 ($\pm$ 0.17) on our liver CT dataset. It also set new record FIDs of 10.78, 3.52, 21.17, and 5.39 on the publicly available SLIVER07, ChestX-ray14, ACDC, and Medical Segmentation Decathlon (brain tumors) datasets. In the visual Turing test, the clinicians rated generated images as real 42% of the time, approaching random guessing. Our computational ablation study revealed that transfer learning and data augmentation stabilize training and improve the perceptual quality of the generated images. We observed the FID to be consistent with human perceptual evaluation of medical images. Finally, our work found that StyleGAN2-ADA consistently produces high-quality results without hyperparameter searches or retraining.
* This preprint has not undergone post-submission improvements or corrections. The Version of Record of this contribution is published in LNCS, volume 13570, and is available online at https://doi.org/10.1007/978-3-031-16980-9_14
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge