Alexander Shieh
The University of Texas MD Anderson Cancer Center
Correcting Class Imbalances with Self-Training for Improved Universal Lesion Detection and Tagging
Apr 07, 2025Abstract:Universal lesion detection and tagging (ULDT) in CT studies is critical for tumor burden assessment and tracking the progression of lesion status (growth/shrinkage) over time. However, a lack of fully annotated data hinders the development of effective ULDT approaches. Prior work used the DeepLesion dataset (4,427 patients, 10,594 studies, 32,120 CT slices, 32,735 lesions, 8 body part labels) for algorithmic development, but this dataset is not completely annotated and contains class imbalances. To address these issues, in this work, we developed a self-training pipeline for ULDT. A VFNet model was trained on a limited 11.5\% subset of DeepLesion (bounding boxes + tags) to detect and classify lesions in CT studies. Then, it identified and incorporated novel lesion candidates from a larger unseen data subset into its training set, and self-trained itself over multiple rounds. Multiple self-training experiments were conducted with different threshold policies to select predicted lesions with higher quality and cover the class imbalances. We discovered that direct self-training improved the sensitivities of over-represented lesion classes at the expense of under-represented classes. However, upsampling the lesions mined during self-training along with a variable threshold policy yielded a 6.5\% increase in sensitivity at 4 FP in contrast to self-training without class balancing (72\% vs 78.5\%) and a 11.7\% increase compared to the same self-training policy without upsampling (66.8\% vs 78.5\%). Furthermore, we show that our results either improved or maintained the sensitivity at 4FP for all 8 lesion classes.
Weakly-Supervised Detection of Bone Lesions in CT
Jan 31, 2024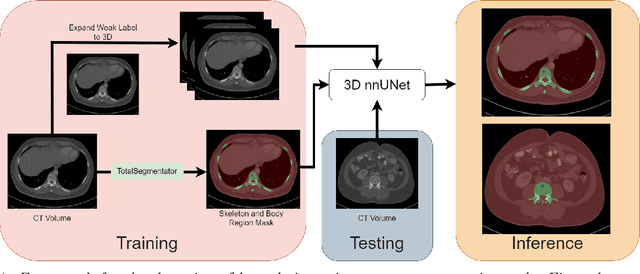
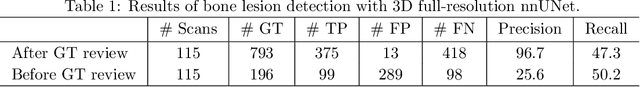
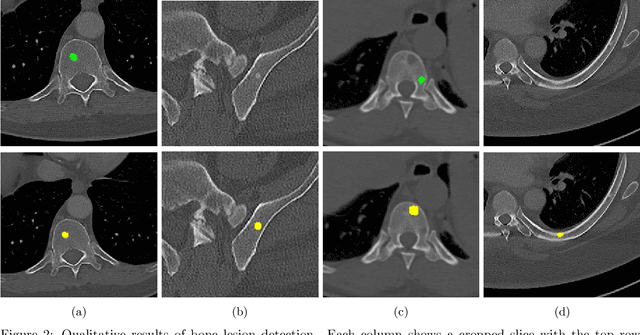
Abstract:The skeletal region is one of the common sites of metastatic spread of cancer in the breast and prostate. CT is routinely used to measure the size of lesions in the bones. However, they can be difficult to spot due to the wide variations in their sizes, shapes, and appearances. Precise localization of such lesions would enable reliable tracking of interval changes (growth, shrinkage, or unchanged status). To that end, an automated technique to detect bone lesions is highly desirable. In this pilot work, we developed a pipeline to detect bone lesions (lytic, blastic, and mixed) in CT volumes via a proxy segmentation task. First, we used the bone lesions that were prospectively marked by radiologists in a few 2D slices of CT volumes and converted them into weak 3D segmentation masks. Then, we trained a 3D full-resolution nnUNet model using these weak 3D annotations to segment the lesions and thereby detected them. Our automated method detected bone lesions in CT with a precision of 96.7% and recall of 47.3% despite the use of incomplete and partial training data. To the best of our knowledge, we are the first to attempt the direct detection of bone lesions in CT via a proxy segmentation task.
Importance of Feature Extraction in the Calculation of Fréchet Distance for Medical Imaging
Nov 22, 2023Abstract:Fr\'echet Inception Distance is a widely used metric for evaluating synthetic image quality that utilizes an ImageNet-trained InceptionV3 network as a feature extractor. However, its application in medical imaging lacks a standard feature extractor, leading to biased and inconsistent comparisons. This study aimed to compare state-of-the-art feature extractors for computing Fr\'echet Distances (FDs) in medical imaging. A StyleGAN2 network was trained with data augmentation techniques tailored for limited data domains on datasets comprising three medical imaging modalities and four anatomical locations. Human evaluation of generative quality (via a visual Turing test) was compared to FDs calculated using ImageNet-trained InceptionV3, ResNet50, SwAV, DINO, and Swin Transformer architectures, in addition to an InceptionV3 network trained on a large medical dataset, RadImageNet. All ImageNet-based extractors were consistent with each other, but only SwAV was significantly correlated with medical expert judgment. The RadImageNet-based FD showed volatility and lacked correlation with human judgment. Caution is advised when using medical image-trained extraction networks in the FD calculation. These networks should be rigorously evaluated on the imaging modality under consideration and publicly released. ImageNet-based extractors, while imperfect, are consistent and widely understood. Training extraction networks with SwAV is a promising approach for synthetic medical image evaluation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge