Angshuman Paul
3D Universal Lesion Detection and Tagging in CT with Self-Training
Apr 07, 2025Abstract:Radiologists routinely perform the tedious task of lesion localization, classification, and size measurement in computed tomography (CT) studies. Universal lesion detection and tagging (ULDT) can simultaneously help alleviate the cumbersome nature of lesion measurement and enable tumor burden assessment. Previous ULDT approaches utilize the publicly available DeepLesion dataset, however it does not provide the full volumetric (3D) extent of lesions and also displays a severe class imbalance. In this work, we propose a self-training pipeline to detect 3D lesions and tag them according to the body part they occur in. We used a significantly limited 30\% subset of DeepLesion to train a VFNet model for 2D lesion detection and tagging. Next, the 2D lesion context was expanded into 3D, and the mined 3D lesion proposals were integrated back into the baseline training data in order to retrain the model over multiple rounds. Through the self-training procedure, our VFNet model learned from its own predictions, detected lesions in 3D, and tagged them. Our results indicated that our VFNet model achieved an average sensitivity of 46.9\% at [0.125:8] false positives (FP) with a limited 30\% data subset in comparison to the 46.8\% of an existing approach that used the entire DeepLesion dataset. To our knowledge, we are the first to jointly detect lesions in 3D and tag them according to the body part label.
Correcting Class Imbalances with Self-Training for Improved Universal Lesion Detection and Tagging
Apr 07, 2025Abstract:Universal lesion detection and tagging (ULDT) in CT studies is critical for tumor burden assessment and tracking the progression of lesion status (growth/shrinkage) over time. However, a lack of fully annotated data hinders the development of effective ULDT approaches. Prior work used the DeepLesion dataset (4,427 patients, 10,594 studies, 32,120 CT slices, 32,735 lesions, 8 body part labels) for algorithmic development, but this dataset is not completely annotated and contains class imbalances. To address these issues, in this work, we developed a self-training pipeline for ULDT. A VFNet model was trained on a limited 11.5\% subset of DeepLesion (bounding boxes + tags) to detect and classify lesions in CT studies. Then, it identified and incorporated novel lesion candidates from a larger unseen data subset into its training set, and self-trained itself over multiple rounds. Multiple self-training experiments were conducted with different threshold policies to select predicted lesions with higher quality and cover the class imbalances. We discovered that direct self-training improved the sensitivities of over-represented lesion classes at the expense of under-represented classes. However, upsampling the lesions mined during self-training along with a variable threshold policy yielded a 6.5\% increase in sensitivity at 4 FP in contrast to self-training without class balancing (72\% vs 78.5\%) and a 11.7\% increase compared to the same self-training policy without upsampling (66.8\% vs 78.5\%). Furthermore, we show that our results either improved or maintained the sensitivity at 4FP for all 8 lesion classes.
Few-shot Diagnosis of Chest x-rays Using an Ensemble of Random Discriminative Subspaces
Aug 31, 2023Abstract:Due to the scarcity of annotated data in the medical domain, few-shot learning may be useful for medical image analysis tasks. We design a few-shot learning method using an ensemble of random subspaces for the diagnosis of chest x-rays (CXRs). Our design is computationally efficient and almost 1.8 times faster than method that uses the popular truncated singular value decomposition (t-SVD) for subspace decomposition. The proposed method is trained by minimizing a novel loss function that helps create well-separated clusters of training data in discriminative subspaces. As a result, minimizing the loss maximizes the distance between the subspaces, making them discriminative and assisting in better classification. Experiments on large-scale publicly available CXR datasets yield promising results. Code for the project will be available at https://github.com/Few-shot-Learning-on-chest-x-ray/fsl_subspace.
Gland Segmentation in Histopathology Images Using Random Forest Guided Boundary Construction
Aug 15, 2017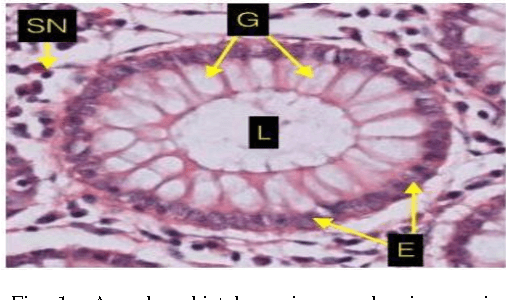
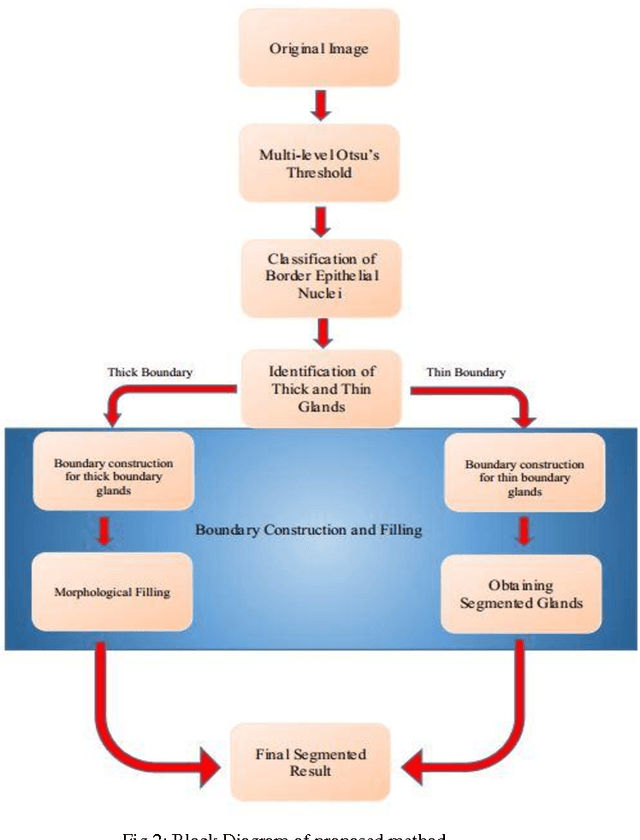
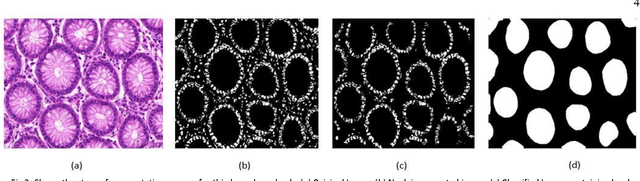
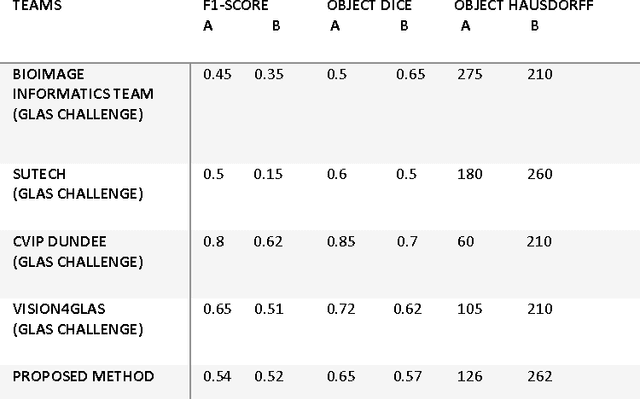
Abstract:Grading of cancer is important to know the extent of its spread. Prior to grading, segmentation of glandular structures is important. Manual segmentation is a time consuming process and is subject to observer bias. Hence, an automated process is required to segment the gland structures. These glands show a large variation in shape size and texture. This makes the task challenging as the glands cannot be segmented using mere morphological operations and conventional segmentation mechanisms. In this project we propose a method which detects the boundary epithelial cells of glands and then a novel approach is used to construct the complete gland boundary. The region enclosed within the boundary can then be obtained to get the segmented gland regions.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge