Rohith AP
Deep Discriminative Learning for Unsupervised Domain Adaptation
Nov 17, 2018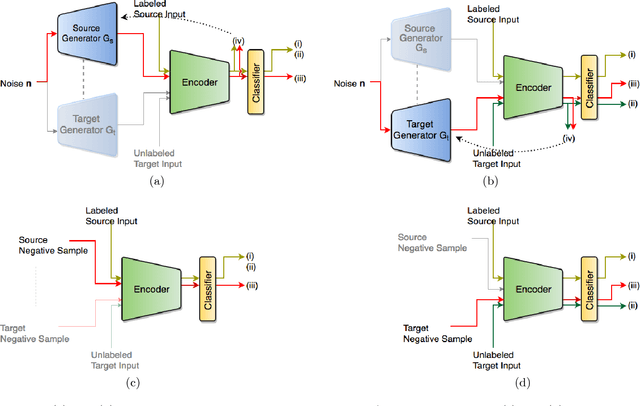
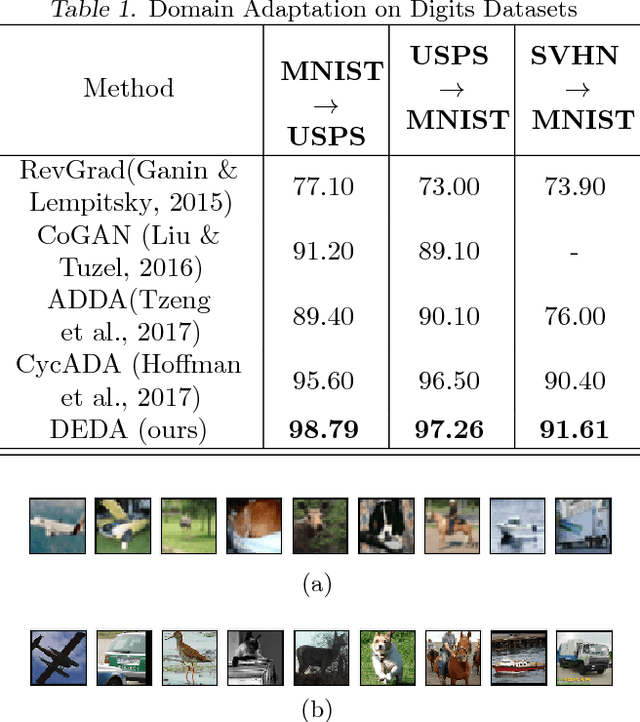

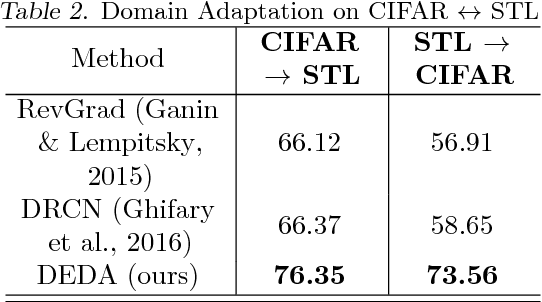
Abstract:The primary objective of domain adaptation methods is to transfer knowledge from a source domain to a target domain that has similar but different data distributions. Thus, in order to correctly classify the unlabeled target domain samples, the standard approach is to learn a common representation for both source and target domain, thereby indirectly addressing the problem of learning a classifier in the target domain. However, such an approach does not address the task of classification in the target domain directly. In contrast, we propose an approach that directly addresses the problem of learning a classifier in the unlabeled target domain. In particular, we train a classifier to correctly classify the training samples while simultaneously classifying the samples in the target domain in an unsupervised manner. The corresponding model is referred to as Discriminative Encoding for Domain Adaptation (DEDA). We show that this simple approach for performing unsupervised domain adaptation is indeed quite powerful. Our method achieves state of the art results in unsupervised adaptation tasks on various image classification benchmarks. We also obtained state of the art performance on domain adaptation in Amazon reviews sentiment classification dataset. We perform additional experiments when the source data has less labeled examples and also on zero-shot domain adaptation task where no target domain samples are used for training.
Gland Segmentation in Histopathology Images Using Random Forest Guided Boundary Construction
Aug 15, 2017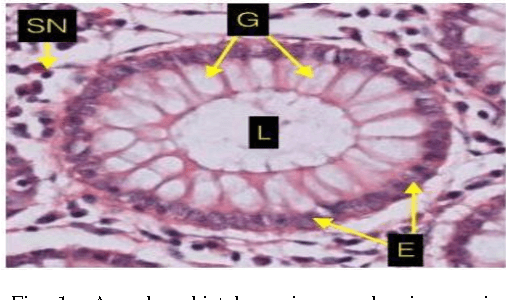
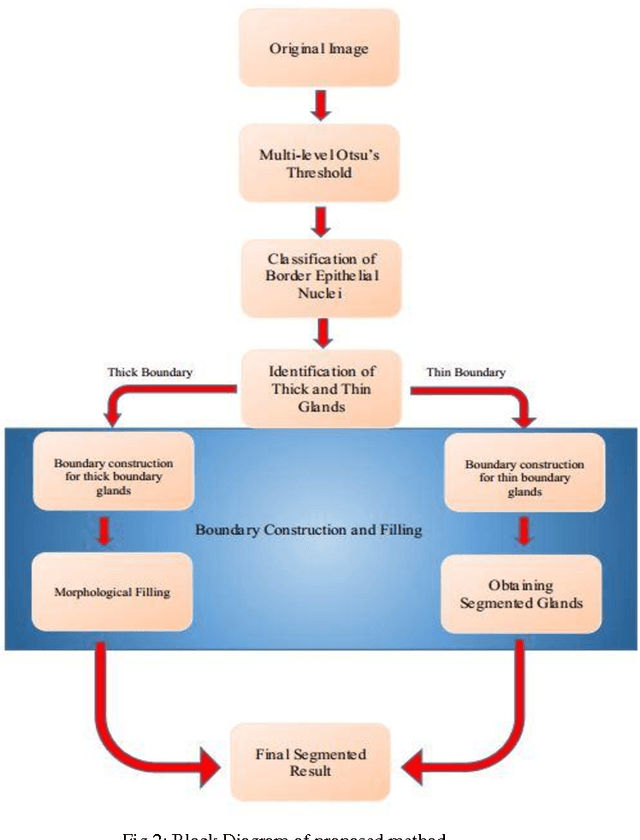
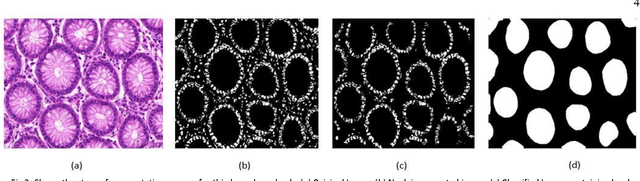
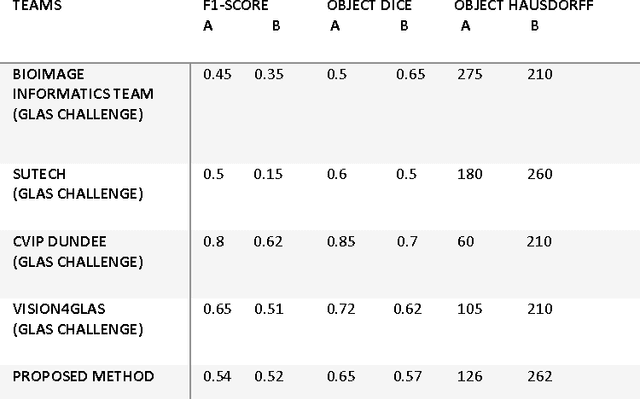
Abstract:Grading of cancer is important to know the extent of its spread. Prior to grading, segmentation of glandular structures is important. Manual segmentation is a time consuming process and is subject to observer bias. Hence, an automated process is required to segment the gland structures. These glands show a large variation in shape size and texture. This makes the task challenging as the glands cannot be segmented using mere morphological operations and conventional segmentation mechanisms. In this project we propose a method which detects the boundary epithelial cells of glands and then a novel approach is used to construct the complete gland boundary. The region enclosed within the boundary can then be obtained to get the segmented gland regions.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge