Kevin McLoughlin
High-Throughput Virtual Screening of Small Molecule Inhibitors for SARS-CoV-2 Protein Targets with Deep Fusion Models
Apr 09, 2021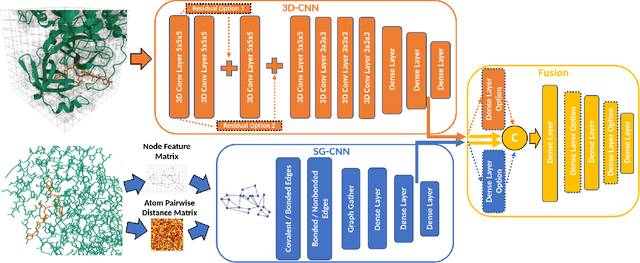
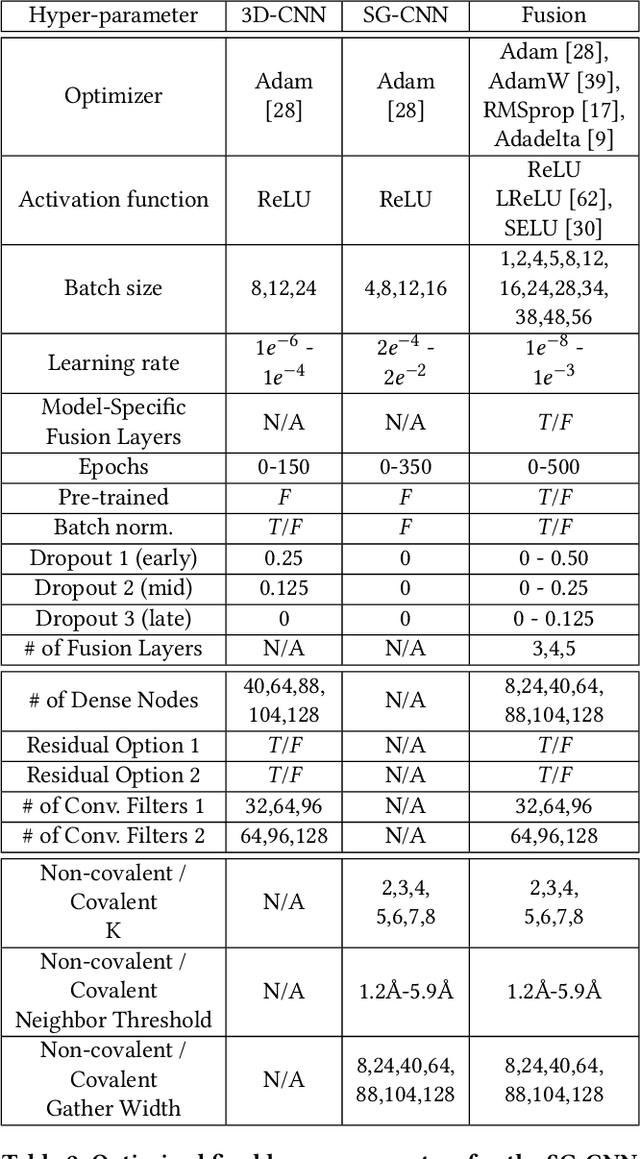
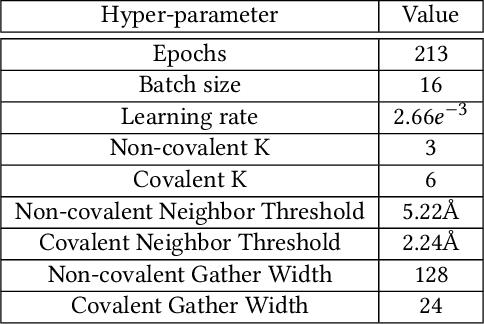
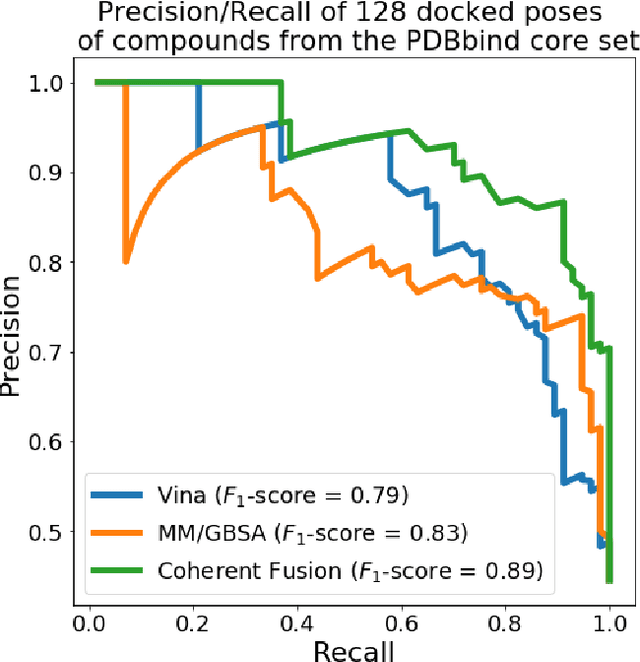
Abstract:Structure-based Deep Fusion models were recently shown to outperform several physics- and machine learning-based protein-ligand binding affinity prediction methods. As part of a multi-institutional COVID-19 pandemic response, over 500 million small molecules were computationally screened against four protein structures from the novel coronavirus (SARS-CoV-2), which causes COVID-19. Three enhancements to Deep Fusion were made in order to evaluate more than 5 billion docked poses on SARS-CoV-2 protein targets. First, the Deep Fusion concept was refined by formulating the architecture as one, coherently backpropagated model (Coherent Fusion) to improve binding-affinity prediction accuracy. Secondly, the model was trained using a distributed, genetic hyper-parameter optimization. Finally, a scalable, high-throughput screening capability was developed to maximize the number of ligands evaluated and expedite the path to experimental evaluation. In this work, we present both the methods developed for machine learning-based high-throughput screening and results from using our computational pipeline to find SARS-CoV-2 inhibitors.
AMPL: A Data-Driven Modeling Pipeline for Drug Discovery
Nov 14, 2019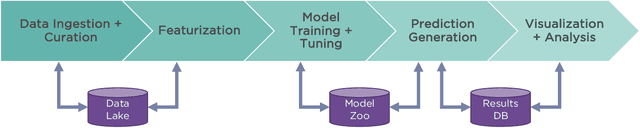

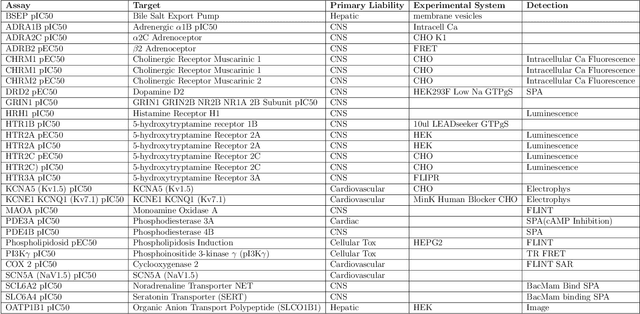
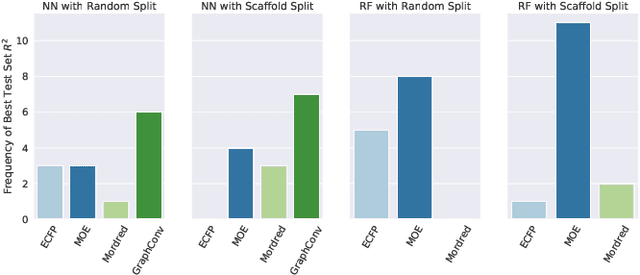
Abstract:One of the key requirements for incorporating machine learning into the drug discovery process is complete reproducibility and traceability of the model building and evaluation process. With this in mind, we have developed an end-to-end modular and extensible software pipeline for building and sharing machine learning models that predict key pharma-relevant parameters. The ATOM Modeling PipeLine, or AMPL, extends the functionality of the open source library DeepChem and supports an array of machine learning and molecular featurization tools. We have benchmarked AMPL on a large collection of pharmaceutical datasets covering a wide range of parameters. As a result of these comprehensive experiments, we have found that physicochemical descriptors and deep learning-based graph representations significantly outperform traditional fingerprints in the characterization of molecular features. We have also found that dataset size is directly correlated to prediction performance, and that single-task deep learning models only outperform shallow learners if there is sufficient data. Likewise, dataset size has a direct impact on model predictivity, independent of comprehensive hyperparameter model tuning. Our findings point to the need for public dataset integration or multi-task/transfer learning approaches. Lastly, we found that uncertainty quantification (UQ) analysis may help identify model error; however, efficacy of UQ to filter predictions varies considerably between datasets and featurization/model types. AMPL is open source and available for download at http://github.com/ATOMconsortium/AMPL.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge