Katikapalli Subramanyam Kalyan
A Survey of GPT-3 Family Large Language Models Including ChatGPT and GPT-4
Oct 04, 2023Abstract:Large language models (LLMs) are a special class of pretrained language models obtained by scaling model size, pretraining corpus and computation. LLMs, because of their large size and pretraining on large volumes of text data, exhibit special abilities which allow them to achieve remarkable performances without any task-specific training in many of the natural language processing tasks. The era of LLMs started with OpenAI GPT-3 model, and the popularity of LLMs is increasing exponentially after the introduction of models like ChatGPT and GPT4. We refer to GPT-3 and its successor OpenAI models, including ChatGPT and GPT4, as GPT-3 family large language models (GLLMs). With the ever-rising popularity of GLLMs, especially in the research community, there is a strong need for a comprehensive survey which summarizes the recent research progress in multiple dimensions and can guide the research community with insightful future research directions. We start the survey paper with foundation concepts like transformers, transfer learning, self-supervised learning, pretrained language models and large language models. We then present a brief overview of GLLMs and discuss the performances of GLLMs in various downstream tasks, specific domains and multiple languages. We also discuss the data labelling and data augmentation abilities of GLLMs, the robustness of GLLMs, the effectiveness of GLLMs as evaluators, and finally, conclude with multiple insightful future research directions. To summarize, this comprehensive survey paper will serve as a good resource for both academic and industry people to stay updated with the latest research related to GPT-3 family large language models.
AMMUS : A Survey of Transformer-based Pretrained Models in Natural Language Processing
Aug 28, 2021



Abstract:Transformer-based pretrained language models (T-PTLMs) have achieved great success in almost every NLP task. The evolution of these models started with GPT and BERT. These models are built on the top of transformers, self-supervised learning and transfer learning. Transformed-based PTLMs learn universal language representations from large volumes of text data using self-supervised learning and transfer this knowledge to downstream tasks. These models provide good background knowledge to downstream tasks which avoids training of downstream models from scratch. In this comprehensive survey paper, we initially give a brief overview of self-supervised learning. Next, we explain various core concepts like pretraining, pretraining methods, pretraining tasks, embeddings and downstream adaptation methods. Next, we present a new taxonomy of T-PTLMs and then give brief overview of various benchmarks including both intrinsic and extrinsic. We present a summary of various useful libraries to work with T-PTLMs. Finally, we highlight some of the future research directions which will further improve these models. We strongly believe that this comprehensive survey paper will serve as a good reference to learn the core concepts as well as to stay updated with the recent happenings in T-PTLMs.
AMMU -- A Survey of Transformer-based Biomedical Pretrained Language Models
Apr 16, 2021



Abstract:Transformer-based pretrained language models (PLMs) have started a new era in modern natural language processing (NLP). These models combine the power of transformers, transfer learning, and self-supervised learning (SSL). Following the success of these models in the general domain, the biomedical research community has developed various in-domain PLMs starting from BioBERT to the latest BioMegatron and CoderBERT models. We strongly believe there is a need for a survey paper that can provide a comprehensive survey of various transformer-based biomedical pretrained language models (BPLMs). In this survey, we start with a brief overview of foundational concepts like self-supervised learning, embedding layer and transformer encoder layers. We discuss core concepts of transformer-based PLMs like pretraining methods, pretraining tasks, fine-tuning methods, and various embedding types specific to biomedical domain. We introduce a taxonomy for transformer-based BPLMs and then discuss all the models. We discuss various challenges and present possible solutions. We conclude by highlighting some of the open issues which will drive the research community to further improve transformer-based BPLMs.
A Hybrid Approach to Measure Semantic Relatedness in Biomedical Concepts
Jan 25, 2021
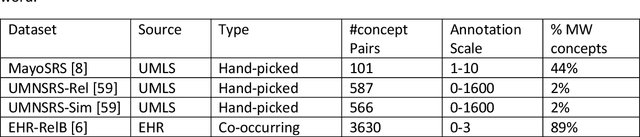
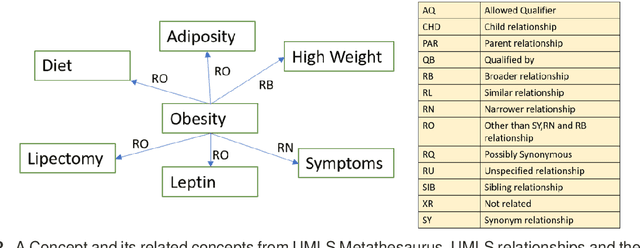

Abstract:Objective: This work aimed to demonstrate the effectiveness of a hybrid approach based on Sentence BERT model and retrofitting algorithm to compute relatedness between any two biomedical concepts. Materials and Methods: We generated concept vectors by encoding concept preferred terms using ELMo, BERT, and Sentence BERT models. We used BioELMo and Clinical ELMo. We used Ontology Knowledge Free (OKF) models like PubMedBERT, BioBERT, BioClinicalBERT, and Ontology Knowledge Injected (OKI) models like SapBERT, CoderBERT, KbBERT, and UmlsBERT. We trained all the BERT models using Siamese network on SNLI and STSb datasets to allow the models to learn more semantic information at the phrase or sentence level so that they can represent multi-word concepts better. Finally, to inject ontology relationship knowledge into concept vectors, we used retrofitting algorithm and concepts from various UMLS relationships. We evaluated our hybrid approach on four publicly available datasets which also includes the recently released EHR-RelB dataset. EHR-RelB is the largest publicly available relatedness dataset in which 89% of terms are multi-word which makes it more challenging. Results: Sentence BERT models mostly outperformed corresponding BERT models. The concept vectors generated using the Sentence BERT model based on SapBERT and retrofitted using UMLS-related concepts achieved the best results on all four datasets. Conclusions: Sentence BERT models are more effective compared to BERT models in computing relatedness scores in most of the cases. Injecting ontology knowledge into concept vectors further enhances their quality and contributes to better relatedness scores.
Want to Identify, Extract and Normalize Adverse Drug Reactions in Tweets? Use RoBERTa
Jun 29, 2020
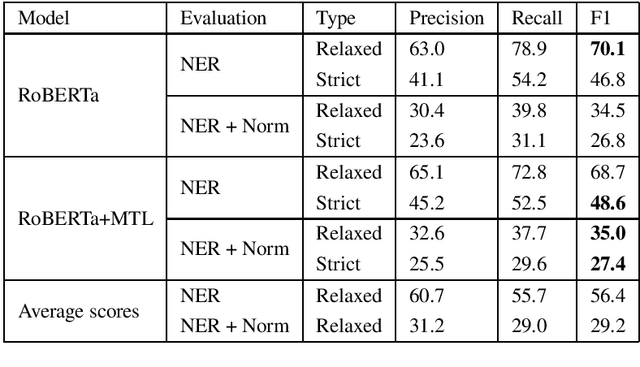
Abstract:This paper presents our approach for task 2 and task 3 of Social Media Mining for Health (SMM4H) 2020 shared tasks. In task 2, we have to differentiate adverse drug reaction (ADR) tweets from nonADR tweets and is treated as binary classification. Task3 involves extracting ADR mentions and then mapping them to MedDRA codes. Extracting ADR mentions is treated as sequence labeling and normalizing ADR mentions is treated as multi-class classification. Our system is based on pre-trained language model RoBERTa and it achieves a) F1-score of 58% in task2 which is 12% more than the average score b) relaxed F1-score of 70.1% in ADR extraction of task 3 which is 13.7% more than the average score and relaxed F1-score of 35% in ADR extraction + normalization of task3 which is 5.8% more than the average score. Overall, our models achieve promising results in both the tasks with significant improvements over average scores.
Medical Concept Normalization in User Generated Texts by Learning Target Concept Embeddings
Jun 07, 2020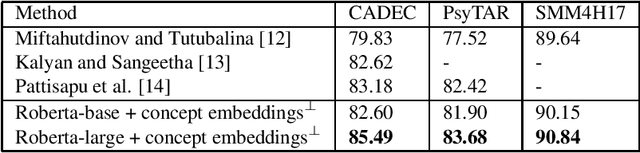
Abstract:Medical concept normalization helps in discovering standard concepts in free-form text i.e., maps health-related mentions to standard concepts in a vocabulary. It is much beyond simple string matching and requires a deep semantic understanding of concept mentions. Recent research approach concept normalization as either text classification or text matching. The main drawback in existing a) text classification approaches is ignoring valuable target concepts information in learning input concept mention representation b) text matching approach is the need to separately generate target concept embeddings which is time and resource consuming. Our proposed model overcomes these drawbacks by jointly learning the representations of input concept mention and target concepts. First, it learns the input concept mention representation using RoBERTa. Second, it finds cosine similarity between embeddings of input concept mention and all the target concepts. Here, embeddings of target concepts are randomly initialized and then updated during training. Finally, the target concept with maximum cosine similarity is assigned to the input concept mention. Our model surpasses all the existing methods across three standard datasets by improving accuracy up to 2.31%.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge