Sivanesan Sangeetha
AMMUS : A Survey of Transformer-based Pretrained Models in Natural Language Processing
Aug 28, 2021



Abstract:Transformer-based pretrained language models (T-PTLMs) have achieved great success in almost every NLP task. The evolution of these models started with GPT and BERT. These models are built on the top of transformers, self-supervised learning and transfer learning. Transformed-based PTLMs learn universal language representations from large volumes of text data using self-supervised learning and transfer this knowledge to downstream tasks. These models provide good background knowledge to downstream tasks which avoids training of downstream models from scratch. In this comprehensive survey paper, we initially give a brief overview of self-supervised learning. Next, we explain various core concepts like pretraining, pretraining methods, pretraining tasks, embeddings and downstream adaptation methods. Next, we present a new taxonomy of T-PTLMs and then give brief overview of various benchmarks including both intrinsic and extrinsic. We present a summary of various useful libraries to work with T-PTLMs. Finally, we highlight some of the future research directions which will further improve these models. We strongly believe that this comprehensive survey paper will serve as a good reference to learn the core concepts as well as to stay updated with the recent happenings in T-PTLMs.
AMMU -- A Survey of Transformer-based Biomedical Pretrained Language Models
Apr 16, 2021



Abstract:Transformer-based pretrained language models (PLMs) have started a new era in modern natural language processing (NLP). These models combine the power of transformers, transfer learning, and self-supervised learning (SSL). Following the success of these models in the general domain, the biomedical research community has developed various in-domain PLMs starting from BioBERT to the latest BioMegatron and CoderBERT models. We strongly believe there is a need for a survey paper that can provide a comprehensive survey of various transformer-based biomedical pretrained language models (BPLMs). In this survey, we start with a brief overview of foundational concepts like self-supervised learning, embedding layer and transformer encoder layers. We discuss core concepts of transformer-based PLMs like pretraining methods, pretraining tasks, fine-tuning methods, and various embedding types specific to biomedical domain. We introduce a taxonomy for transformer-based BPLMs and then discuss all the models. We discuss various challenges and present possible solutions. We conclude by highlighting some of the open issues which will drive the research community to further improve transformer-based BPLMs.
A Hybrid Approach to Measure Semantic Relatedness in Biomedical Concepts
Jan 25, 2021
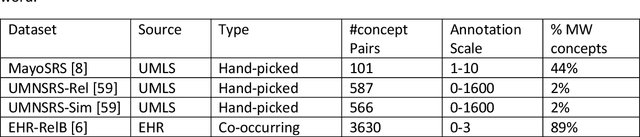
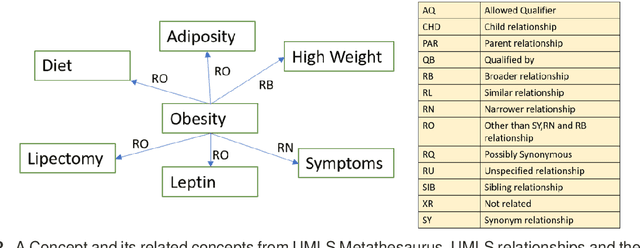

Abstract:Objective: This work aimed to demonstrate the effectiveness of a hybrid approach based on Sentence BERT model and retrofitting algorithm to compute relatedness between any two biomedical concepts. Materials and Methods: We generated concept vectors by encoding concept preferred terms using ELMo, BERT, and Sentence BERT models. We used BioELMo and Clinical ELMo. We used Ontology Knowledge Free (OKF) models like PubMedBERT, BioBERT, BioClinicalBERT, and Ontology Knowledge Injected (OKI) models like SapBERT, CoderBERT, KbBERT, and UmlsBERT. We trained all the BERT models using Siamese network on SNLI and STSb datasets to allow the models to learn more semantic information at the phrase or sentence level so that they can represent multi-word concepts better. Finally, to inject ontology relationship knowledge into concept vectors, we used retrofitting algorithm and concepts from various UMLS relationships. We evaluated our hybrid approach on four publicly available datasets which also includes the recently released EHR-RelB dataset. EHR-RelB is the largest publicly available relatedness dataset in which 89% of terms are multi-word which makes it more challenging. Results: Sentence BERT models mostly outperformed corresponding BERT models. The concept vectors generated using the Sentence BERT model based on SapBERT and retrofitted using UMLS-related concepts achieved the best results on all four datasets. Conclusions: Sentence BERT models are more effective compared to BERT models in computing relatedness scores in most of the cases. Injecting ontology knowledge into concept vectors further enhances their quality and contributes to better relatedness scores.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge