A Hybrid Approach to Measure Semantic Relatedness in Biomedical Concepts
Paper and Code
Jan 25, 2021
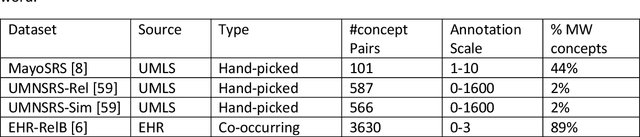
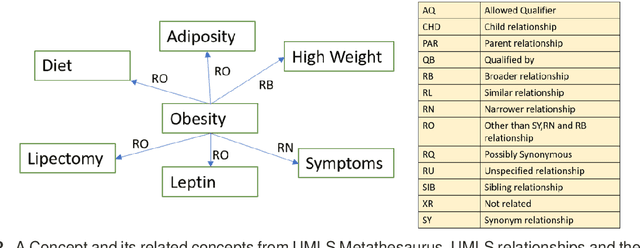

Objective: This work aimed to demonstrate the effectiveness of a hybrid approach based on Sentence BERT model and retrofitting algorithm to compute relatedness between any two biomedical concepts. Materials and Methods: We generated concept vectors by encoding concept preferred terms using ELMo, BERT, and Sentence BERT models. We used BioELMo and Clinical ELMo. We used Ontology Knowledge Free (OKF) models like PubMedBERT, BioBERT, BioClinicalBERT, and Ontology Knowledge Injected (OKI) models like SapBERT, CoderBERT, KbBERT, and UmlsBERT. We trained all the BERT models using Siamese network on SNLI and STSb datasets to allow the models to learn more semantic information at the phrase or sentence level so that they can represent multi-word concepts better. Finally, to inject ontology relationship knowledge into concept vectors, we used retrofitting algorithm and concepts from various UMLS relationships. We evaluated our hybrid approach on four publicly available datasets which also includes the recently released EHR-RelB dataset. EHR-RelB is the largest publicly available relatedness dataset in which 89% of terms are multi-word which makes it more challenging. Results: Sentence BERT models mostly outperformed corresponding BERT models. The concept vectors generated using the Sentence BERT model based on SapBERT and retrofitted using UMLS-related concepts achieved the best results on all four datasets. Conclusions: Sentence BERT models are more effective compared to BERT models in computing relatedness scores in most of the cases. Injecting ontology knowledge into concept vectors further enhances their quality and contributes to better relatedness scores.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge