Kaleem Nawaz Khan
ImageCAS: A Large-Scale Dataset and Benchmark for Coronary Artery Segmentation based on Computed Tomography Angiography Images
Nov 03, 2022Abstract:Cardiovascular disease (CVD) accounts for about half of non-communicable diseases. Vessel stenosis in the coronary artery is considered to be the major risk of CVD. Computed tomography angiography (CTA) is one of the widely used noninvasive imaging modalities in coronary artery diagnosis due to its superior image resolution. Clinically, segmentation of coronary arteries is essential for the diagnosis and quantification of coronary artery disease. Recently, a variety of works have been proposed to address this problem. However, on one hand, most works rely on in-house datasets, and only a few works published their datasets to the public which only contain tens of images. On the other hand, their source code have not been published, and most follow-up works have not made comparison with existing works, which makes it difficult to judge the effectiveness of the methods and hinders the further exploration of this challenging yet critical problem in the community. In this paper, we propose a large-scale dataset for coronary artery segmentation on CTA images. In addition, we have implemented a benchmark in which we have tried our best to implement several typical existing methods. Furthermore, we propose a strong baseline method which combines multi-scale patch fusion and two-stage processing to extract the details of vessels. Comprehensive experiments show that the proposed method achieves better performance than existing works on the proposed large-scale dataset. The benchmark and the dataset are published at https://github.com/XiaoweiXu/ImageCAS-A-Large-Scale-Dataset-and-Benchmark-for-Coronary-Artery-Segmentation-based-on-CT.
Evaluation of Preprocessing Techniques for U-Net Based Automated Liver Segmentation
Mar 26, 2021
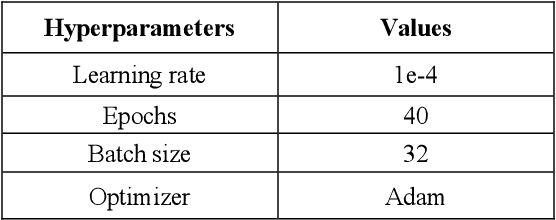
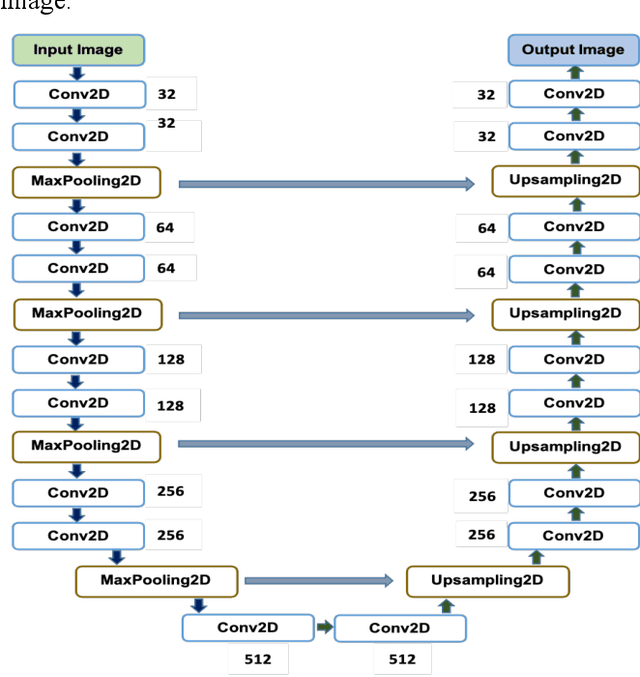
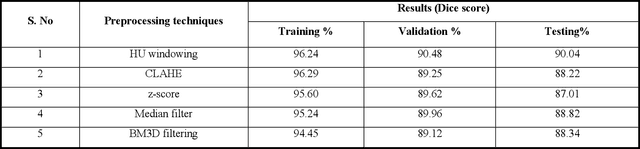
Abstract:To extract liver from medical images is a challenging task due to similar intensity values of liver with adjacent organs, various contrast levels, various noise associated with medical images and irregular shape of liver. To address these issues, it is important to preprocess the medical images, i.e., computerized tomography (CT) and magnetic resonance imaging (MRI) data prior to liver analysis and quantification. This paper investigates the impact of permutation of various preprocessing techniques for CT images, on the automated liver segmentation using deep learning, i.e., U-Net architecture. The study focuses on Hounsfield Unit (HU) windowing, contrast limited adaptive histogram equalization (CLAHE), z-score normalization, median filtering and Block-Matching and 3D (BM3D) filtering. The segmented results show that combination of three techniques; HU-windowing, median filtering and z-score normalization achieve optimal performance with Dice coefficient of 96.93%, 90.77% and 90.84% for training, validation and testing respectively.
Deep Learning Based Classification of Unsegmented Phonocardiogram Spectrograms Leveraging Transfer Learning
Dec 15, 2020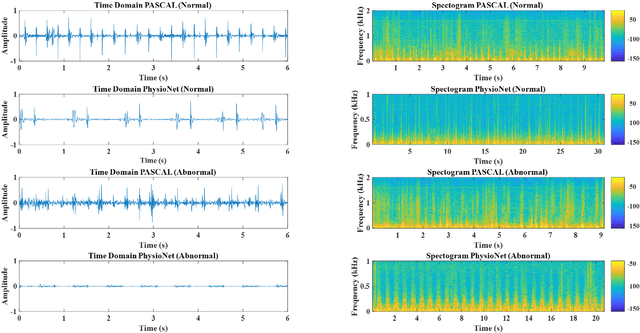
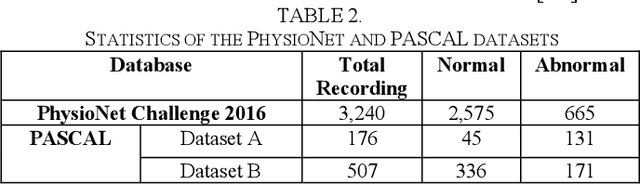
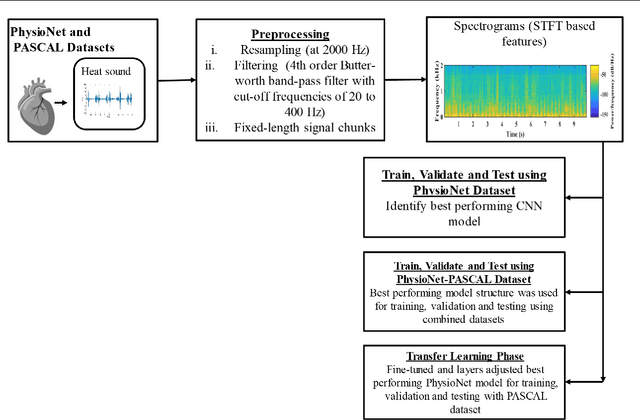
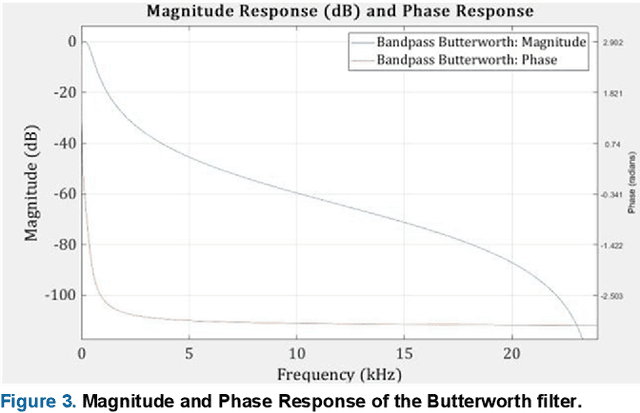
Abstract:Cardiovascular diseases (CVDs) are the main cause of deaths all over the world. Heart murmurs are the most common abnormalities detected during the auscultation process. The two widely used publicly available phonocardiogram (PCG) datasets are from the PhysioNet/CinC (2016) and PASCAL (2011) challenges. The datasets are significantly different in terms of the tools used for data acquisition, clinical protocols, digital storages and signal qualities, making it challenging to process and analyze. In this work, we have used short-time Fourier transform (STFT) based spectrograms to learn the representative patterns of the normal and abnormal PCG signals. Spectrograms generated from both the datasets are utilized to perform three different studies: (i) train, validate and test different variants of convolutional neural network (CNN) models with PhysioNet dataset, (ii) train, validate and test the best performing CNN structure on combined PhysioNet-PASCAL dataset and (iii) finally, transfer learning technique is employed to train the best performing pre-trained network from the first study with PASCAL dataset. We propose a novel, less complex and relatively light custom CNN model for the classification of PhysioNet, combined and PASCAL datasets. The first study achieves an accuracy, sensitivity, specificity, precision and F1 score of 95.4%, 96.3%, 92.4%, 97.6% and 96.98% respectively while the second study shows accuracy, sensitivity, specificity, precision and F1 score of 94.2%, 95.5%, 90.3%, 96.8% and 96.1% respectively. Finally, the third study shows a precision of 98.29% on the noisy PASCAL dataset with transfer learning approach. All the three proposed approaches outperform most of the recent competing studies by achieving comparatively high classification accuracy and precision, which make them suitable for screening CVDs using PCG signals.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge