Zumray Dokur
Classification of Hyperspectral Images by Using Spectral Data and Fully Connected Neural Network
Jan 08, 2022



Abstract:It is observed that high classification performance is achieved for one- and two-dimensional signals by using deep learning methods. In this context, most researchers have tried to classify hyperspectral images by using deep learning methods and classification success over 90% has been achieved for these images. Deep neural networks (DNN) actually consist of two parts: i) Convolutional neural network (CNN) and ii) fully connected neural network (FCNN). While CNN determines the features, FCNN is used in classification. In classification of the hyperspectral images, it is observed that almost all of the researchers used 2D or 3D convolution filters on the spatial data beside spectral data (features). It is convenient to use convolution filters on images or time signals. In hyperspectral images, each pixel is represented by a signature vector which consists of individual features that are independent of each other. Since the order of the features in the vector can be changed, it doesn't make sense to use convolution filters on these features as on time signals. At the same time, since the hyperspectral images do not have a textural structure, there is no need to use spatial data besides spectral data. In this study, hyperspectral images of Indian pines, Salinas, Pavia centre, Pavia university and Botswana are classified by using only fully connected neural network and the spectral data with one dimensional. An average accuracy of 97.5% is achieved for the test sets of all hyperspectral images.
Generating Ten BCI Commands Using Four Simple Motor Imageries
May 30, 2021



Abstract:The brain computer interface (BCI) systems are utilized for transferring information among humans and computers by analyzing electroencephalogram (EEG) recordings.The process of mentally previewing a motor movement without generating the corporal output can be described as motor imagery (MI).In this emerging research field, the number of commands is also limited in relation to the number of MI tasks; in the current literature, mostly two or four commands (classes) are studied. As a solution to this problem, it is recommended to use mental tasks as well as MI tasks. Unfortunately, the use of this approach reduces the classification performance of MI EEG signals. The fMRI analyses show that the resources in the brain associated with the motor imagery can be activated independently. It is assumed that the brain activity induced by the MI of the combination of body parts corresponds to the superposition of the activities generated during each body parts's simple MI. In this study, in order to create more than four BCI commands, we suggest to generate combined MI EEG signals artificially by using left hand, right hand, tongue, and feet motor imageries in pairs. A maximum of ten different BCI commands can be generated by using four motor imageries in pairs.This study aims to achieve high classification performances for BCI commands produced from four motor imageries by implementing a small-sized deep neural network (DNN).The presented method is evaluated on the four-class datasets of BCI Competitions III and IV, and an average classification performance of 81.8% is achieved for ten classes. The above assumption is also validated on a different dataset which consists of simple and combined MI EEG signals acquired in real time. Trained with the artificially generated combined MI EEG signals, DivFE resulted in an average of 76.5% success rate for the combined MI EEG signals acquired in real-time.
Classification of Motor Imagery EEG Signals by Using a Divergence Based Convolutional Neural Network
Mar 19, 2021



Abstract:Deep neural networks (DNNs) are observed to be successful in pattern classification. However, high classification performances of DNNs are related to their large training sets. Unfortunately, in the literature, the datasets used to classify motor imagery (MI) electroencephalogram (EEG) signals contain a small number of samples. To achieve high performances with small-sized datasets, most of the studies have employed a transformation such as common spatial patterns (CSP) before the classification process. However, CSP is dependent on subjects and introduces computational load in real-time applications. It is observed in the literature that the augmentation process is not applied for increasing the classification performance of EEG signals. In this study, we have investigated the effect of the augmentation process on the classification performance of MI EEG signals instead of using a preceding transformation such as the CSP, and we have demonstrated that by resulting in high success rates for the classification of MI EEGs, the augmentation process is able to compete with the CSP. In addition to the augmentation process, we modified the DNN structure to increase the classification performance, to decrease the number of nodes in the structure, and to be used with less number of hyper parameters. A minimum distance network (MDN) following the last layer of the convolutional neural network (CNN) was used as the classifier instead of a fully connected neural network (FCNN). By augmenting the EEG dataset and focusing solely on CNN's training, the training algorithm of the proposed structure is strengthened without applying any transformation. We tested these improvements on brain-computer interface (BCI) competitions 2005 and 2008 databases with two and four classes, and the high impact of the augmentation on the average performances are demonstrated.
Deep Learning Based Classification of Unsegmented Phonocardiogram Spectrograms Leveraging Transfer Learning
Dec 15, 2020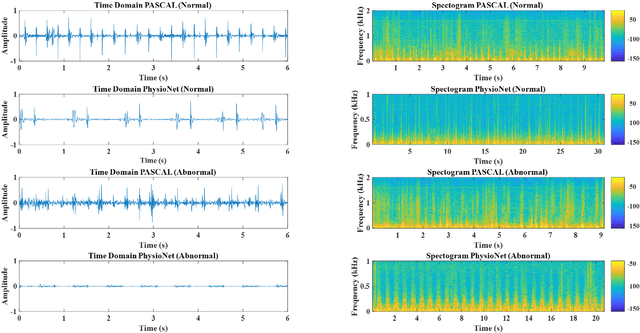
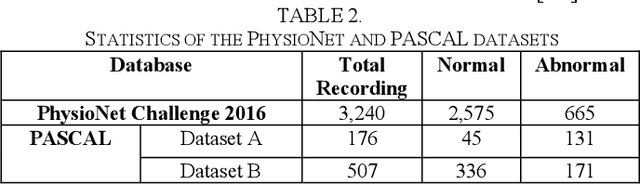
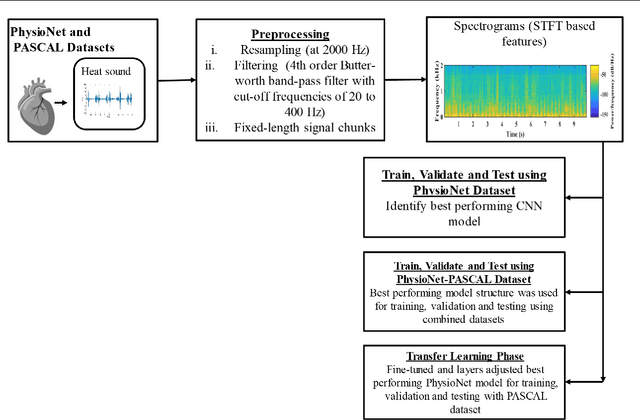
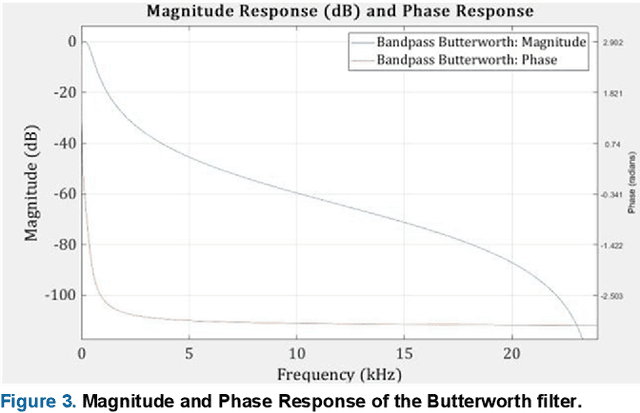
Abstract:Cardiovascular diseases (CVDs) are the main cause of deaths all over the world. Heart murmurs are the most common abnormalities detected during the auscultation process. The two widely used publicly available phonocardiogram (PCG) datasets are from the PhysioNet/CinC (2016) and PASCAL (2011) challenges. The datasets are significantly different in terms of the tools used for data acquisition, clinical protocols, digital storages and signal qualities, making it challenging to process and analyze. In this work, we have used short-time Fourier transform (STFT) based spectrograms to learn the representative patterns of the normal and abnormal PCG signals. Spectrograms generated from both the datasets are utilized to perform three different studies: (i) train, validate and test different variants of convolutional neural network (CNN) models with PhysioNet dataset, (ii) train, validate and test the best performing CNN structure on combined PhysioNet-PASCAL dataset and (iii) finally, transfer learning technique is employed to train the best performing pre-trained network from the first study with PASCAL dataset. We propose a novel, less complex and relatively light custom CNN model for the classification of PhysioNet, combined and PASCAL datasets. The first study achieves an accuracy, sensitivity, specificity, precision and F1 score of 95.4%, 96.3%, 92.4%, 97.6% and 96.98% respectively while the second study shows accuracy, sensitivity, specificity, precision and F1 score of 94.2%, 95.5%, 90.3%, 96.8% and 96.1% respectively. Finally, the third study shows a precision of 98.29% on the noisy PASCAL dataset with transfer learning approach. All the three proposed approaches outperform most of the recent competing studies by achieving comparatively high classification accuracy and precision, which make them suitable for screening CVDs using PCG signals.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge