Josep Malvehy
DermaVQA-DAS: Dermatology Assessment Schema (DAS) & Datasets for Closed-Ended Question Answering & Segmentation in Patient-Generated Dermatology Images
Dec 30, 2025Abstract:Recent advances in dermatological image analysis have been driven by large-scale annotated datasets; however, most existing benchmarks focus on dermatoscopic images and lack patient-authored queries and clinical context, limiting their applicability to patient-centered care. To address this gap, we introduce DermaVQA-DAS, an extension of the DermaVQA dataset that supports two complementary tasks: closed-ended question answering (QA) and dermatological lesion segmentation. Central to this work is the Dermatology Assessment Schema (DAS), a novel expert-developed framework that systematically captures clinically meaningful dermatological features in a structured and standardized form. DAS comprises 36 high-level and 27 fine-grained assessment questions, with multiple-choice options in English and Chinese. Leveraging DAS, we provide expert-annotated datasets for both closed QA and segmentation and benchmark state-of-the-art multimodal models. For segmentation, we evaluate multiple prompting strategies and show that prompt design impacts performance: the default prompt achieves the best results under Mean-of-Max and Mean-of-Mean evaluation aggregation schemes, while an augmented prompt incorporating both patient query title and content yields the highest performance under majority-vote-based microscore evaluation, achieving a Jaccard index of 0.395 and a Dice score of 0.566 with BiomedParse. For closed-ended QA, overall performance is strong across models, with average accuracies ranging from 0.729 to 0.798; o3 achieves the best overall accuracy (0.798), closely followed by GPT-4.1 (0.796), while Gemini-1.5-Pro shows competitive performance within the Gemini family (0.783). We publicly release DermaVQA-DAS, the DAS schema, and evaluation protocols to support and accelerate future research in patient-centered dermatological vision-language modeling (https://osf.io/72rp3).
The iToBoS dataset: skin region images extracted from 3D total body photographs for lesion detection
Jan 30, 2025



Abstract:Artificial intelligence has significantly advanced skin cancer diagnosis by enabling rapid and accurate detection of malignant lesions. In this domain, most publicly available image datasets consist of single, isolated skin lesions positioned at the center of the image. While these lesion-centric datasets have been fundamental for developing diagnostic algorithms, they lack the context of the surrounding skin, which is critical for improving lesion detection. The iToBoS dataset was created to address this challenge. It includes 16,954 images of skin regions from 100 participants, captured using 3D total body photography. Each image roughly corresponds to a $7 \times 9$ cm section of skin with all suspicious lesions annotated using bounding boxes. Additionally, the dataset provides metadata such as anatomical location, age group, and sun damage score for each image. This dataset aims to facilitate training and benchmarking of algorithms, with the goal of enabling early detection of skin cancer and deployment of this technology in non-clinical environments.
A Patient-Centric Dataset of Images and Metadata for Identifying Melanomas Using Clinical Context
Aug 07, 2020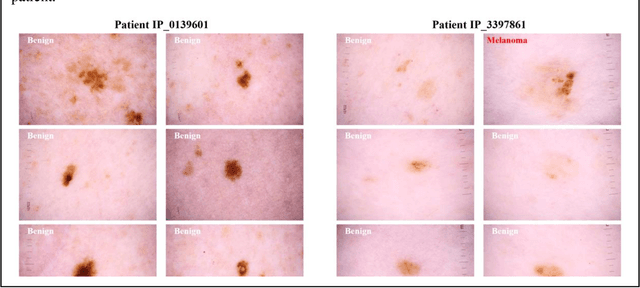
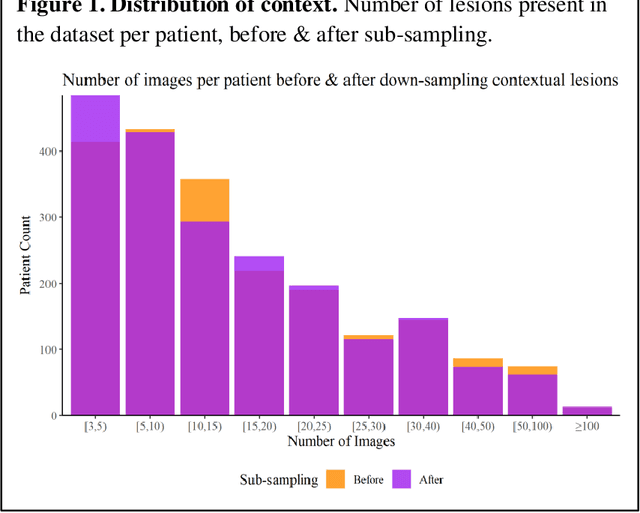
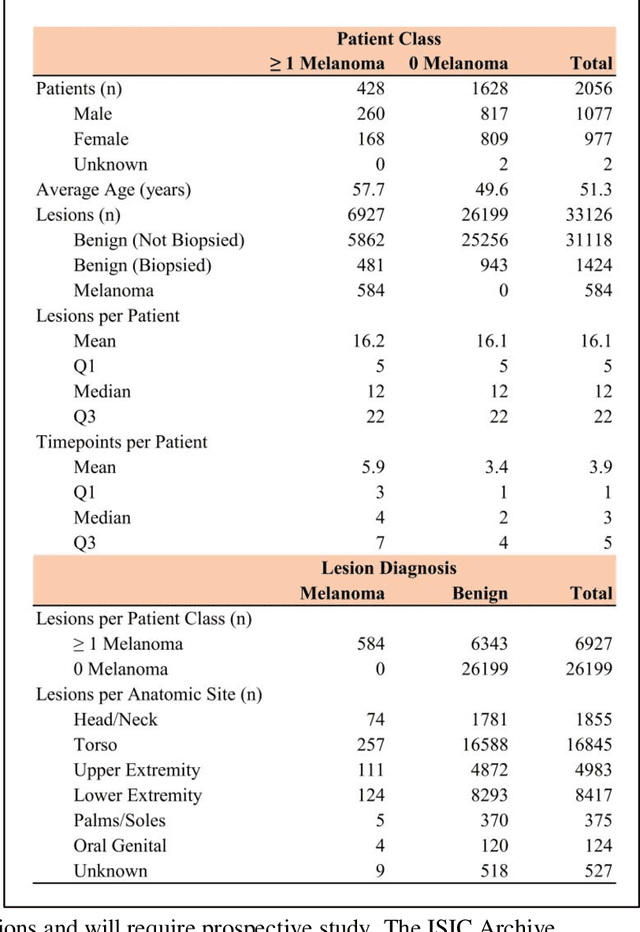
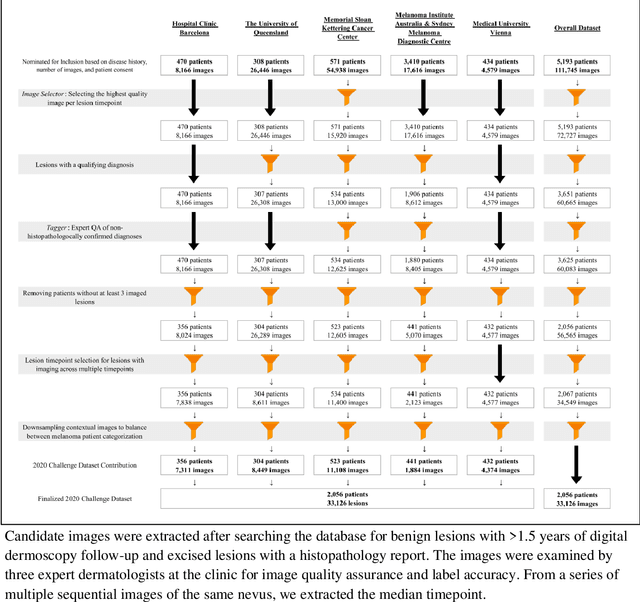
Abstract:Prior skin image datasets have not addressed patient-level information obtained from multiple skin lesions from the same patient. Though artificial intelligence classification algorithms have achieved expert-level performance in controlled studies examining single images, in practice dermatologists base their judgment holistically from multiple lesions on the same patient. The 2020 SIIM-ISIC Melanoma Classification challenge dataset described herein was constructed to address this discrepancy between prior challenges and clinical practice, providing for each image in the dataset an identifier allowing lesions from the same patient to be mapped to one another. This patient-level contextual information is frequently used by clinicians to diagnose melanoma and is especially useful in ruling out false positives in patients with many atypical nevi. The dataset represents 2,056 patients from three continents with an average of 16 lesions per patient, consisting of 33,126 dermoscopic images and 584 histopathologically confirmed melanomas compared with benign melanoma mimickers.
Hi Sigma, do I have the Coronavirus?: Call for a New Artificial Intelligence Approach to Support Health Care Professionals Dealing With The COVID-19 Pandemic
Apr 10, 2020
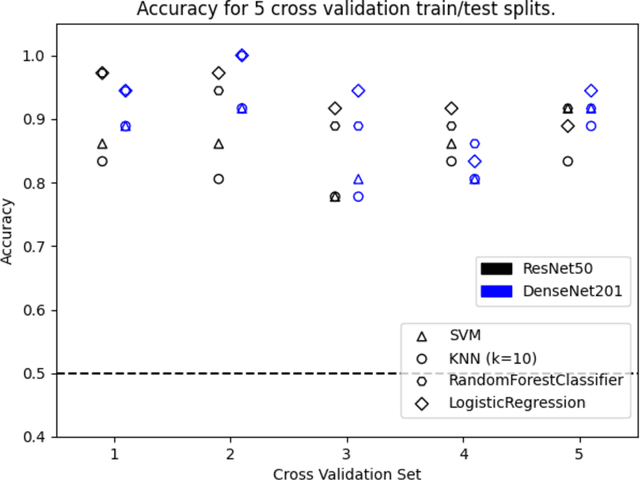

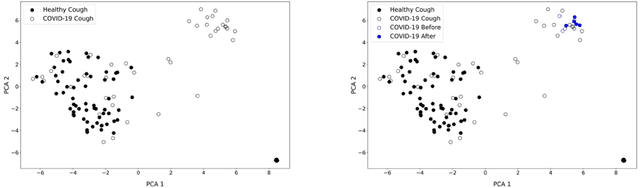
Abstract:Just like your phone can detect what song is playing in crowded spaces, we show that Artificial Intelligence transfer learning algorithms trained on cough phone recordings results in diagnostic tests for COVID-19. To gain adoption by the health care community, we plan to validate our results in a clinical trial and three other venues in Mexico, Spain and the USA . However, if we had data from other on-going clinical trials and volunteers, we may do much more. For example, for confirmed stay-at-home COVID-19 patients, a longitudinal audio test could be developed to determine contact-with-hospital recommendations, and for the most critical COVID-19 patients a success ratio forecast test, including patient clinical data, to prioritize ICU allocation. As a challenge to the engineering community and in the context of our clinical trial, the authors suggest distributing cough recordings daily, hoping other trials and crowdsourcing users will contribute more data. Previous approaches to complex AI tasks have either used a static dataset or were private efforts led by large corporations. All existing COVID-19 trials published also follow this paradigm. Instead, we suggest a novel open collective approach to large-scale real-time health care AI. We will be posting updates at https://opensigma.mit.edu. Our personal view is that our approach is the right one for large scale pandemics, and therefore is here to stay - will you join?
BCN20000: Dermoscopic Lesions in the Wild
Aug 30, 2019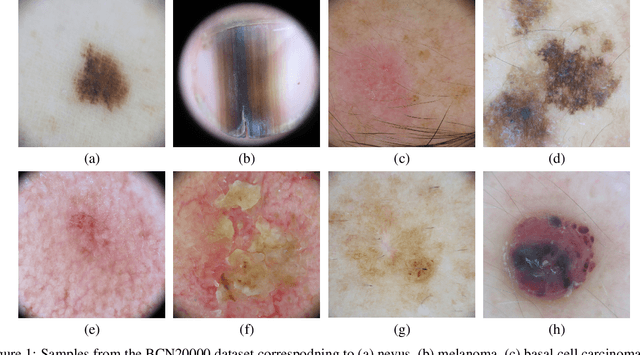
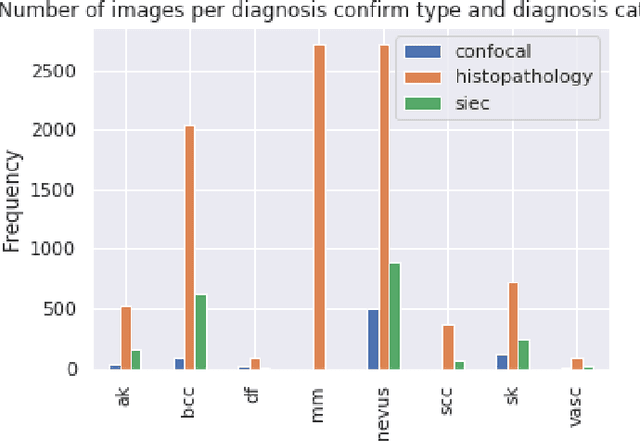
Abstract:This article summarizes the BCN20000 dataset, composed of 19424 dermoscopic images of skin lesions captured from 2010 to 2016 in the facilities of the Hospital Cl\'inic in Barcelona. With this dataset, we aim to study the problem of unconstrained classification of dermoscopic images of skin cancer, including lesions found in hard-to-diagnose locations (nails and mucosa), large lesions which do not fit in the aperture of the dermoscopy device, and hypo-pigmented lesions. The BCN20000 will be provided to the participants of the ISIC Challenge 2019, where they will be asked to train algorithms to classify dermoscopic images of skin cancer automatically.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge