Jorge Henriques
Recovering the Graph Underlying Networked Dynamical Systems under Partial Observability: A Deep Learning Approach
Aug 08, 2022



Abstract:We study the problem of graph structure identification, i.e., of recovering the graph of dependencies among time series. We model these time series data as components of the state of linear stochastic networked dynamical systems. We assume partial observability, where the state evolution of only a subset of nodes comprising the network is observed. We devise a new feature vector computed from the observed time series and prove that these features are linearly separable, i.e., there exists a hyperplane that separates the cluster of features associated with connected pairs of nodes from those associated with disconnected pairs. This renders the features amenable to train a variety of classifiers to perform causal inference. In particular, we use these features to train Convolutional Neural Networks (CNNs). The resulting causal inference mechanism outperforms state-of-the-art counterparts w.r.t. sample-complexity. The trained CNNs generalize well over structurally distinct networks (dense or sparse) and noise-level profiles. Remarkably, they also generalize well to real-world networks while trained over a synthetic network (realization of a random graph). Finally, the proposed method consistently reconstructs the graph in a pairwise manner, that is, by deciding if an edge or arrow is present or absent in each pair of nodes, from the corresponding time series of each pair. This fits the framework of large-scale systems, where observation or processing of all nodes in the network is prohibitive.
A New Approach for Interpretability and Reliability in Clinical Risk Prediction: Acute Coronary Syndrome Scenario
Oct 15, 2021

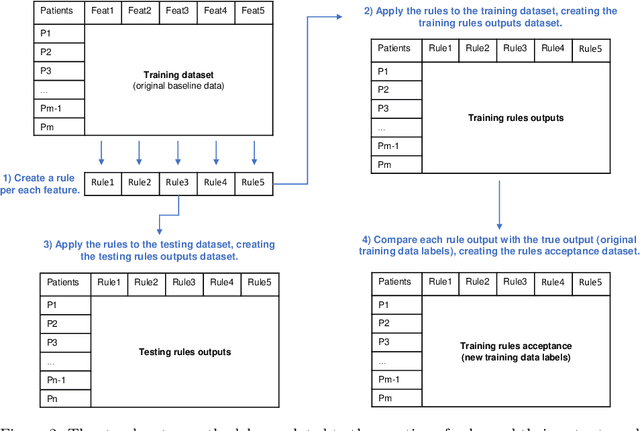
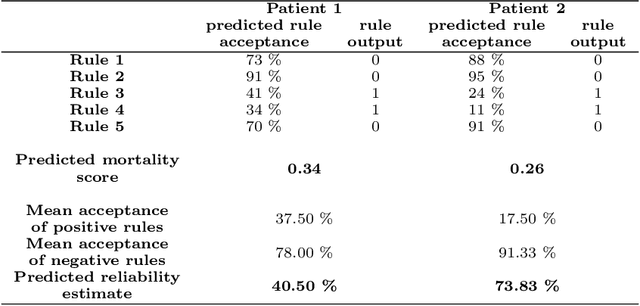
Abstract:We intend to create a new risk assessment methodology that combines the best characteristics of both risk score and machine learning models. More specifically, we aim to develop a method that, besides having a good performance, offers a personalized model and outcome for each patient, presents high interpretability, and incorporates an estimation of the prediction reliability which is not usually available. By combining these features in the same approach we expect that it can boost the confidence of physicians to use such a tool in their daily activity. In order to achieve the mentioned goals, a three-step methodology was developed: several rules were created by dichotomizing risk factors; such rules were trained with a machine learning classifier to predict the acceptance degree of each rule (the probability that the rule is correct) for each patient; that information was combined and used to compute the risk of mortality and the reliability of such prediction. The methodology was applied to a dataset of patients admitted with any type of acute coronary syndromes (ACS), to assess the 30-days all-cause mortality risk. The performance was compared with state-of-the-art approaches: logistic regression (LR), artificial neural network (ANN), and clinical risk score model (Global Registry of Acute Coronary Events - GRACE). The proposed approach achieved testing results identical to the standard LR, but offers superior interpretability and personalization; it also significantly outperforms the GRACE risk model and the standard ANN model. The calibration curve also suggests a very good generalization ability of the obtained model as it approaches the ideal curve. Finally, the reliability estimation of individual predictions presented a great correlation with the misclassifications rate. Those properties may have a beneficial application in other clinical scenarios as well. [abridged]
* Accepted for publication in the Artificial Intelligence in Medicine journal. Abstract abridged to respect the arXiv's characters limit
Personalized and Reliable Decision Sets: Enhancing Interpretability in Clinical Decision Support Systems
Jul 15, 2021



Abstract:In this study, we present a novel clinical decision support system and discuss its interpretability-related properties. It combines a decision set of rules with a machine learning scheme to offer global and local interpretability. More specifically, machine learning is used to predict the likelihood of each of those rules to be correct for a particular patient, which may also contribute to better predictive performances. Moreover, the reliability analysis of individual predictions is also addressed, contributing to further personalized interpretability. The combination of these several elements may be crucial to obtain the clinical stakeholders' trust, leading to a better assessment of patients' conditions and improvement of the physicians' decision-making.
Improving the compromise between accuracy, interpretability and personalization of rule-based machine learning in medical problems
Jun 15, 2021



Abstract:One of the key challenges when developing a predictive model is the capability to describe the domain knowledge and the cause-effect relationships in a simple way. Decision rules are a useful and important methodology in this context, justifying their application in several areas, in particular in clinical practice. Several machine-learning classifiers have exploited the advantageous properties of decision rules to build intelligent prediction models, namely decision trees and ensembles of trees (ETs). However, such methodologies usually suffer from a trade-off between interpretability and predictive performance. Some procedures consider a simplification of ETs, using heuristic approaches to select an optimal reduced set of decision rules. In this paper, we introduce a novel step to those methodologies. We create a new component to predict if a given rule will be correct or not for a particular patient, which introduces personalization into the procedure. Furthermore, the validation results using three public clinical datasets show that it also allows to increase the predictive performance of the selected set of rules, improving the mentioned trade-off.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge