Johannes Hagemann
Digital Health & Machine Learning, Hasso Plattner Institute, University of Potsdam, Germany
INTELLECT-3: Technical Report
Dec 18, 2025Abstract:We present INTELLECT-3, a 106B-parameter Mixture-of-Experts model (12B active) trained with large-scale reinforcement learning on our end-to-end RL infrastructure stack. INTELLECT-3 achieves state of the art performance for its size across math, code, science and reasoning benchmarks, outperforming many larger frontier models. We open-source the model together with the full infrastructure stack used to create it, including RL frameworks, complete recipe, and a wide collection of environments, built with the verifiers library, for training and evaluation from our Environments Hub community platform. Built for this effort, we introduce prime-rl, an open framework for large-scale asynchronous reinforcement learning, which scales seamlessly from a single node to thousands of GPUs, and is tailored for agentic RL with first-class support for multi-turn interactions and tool use. Using this stack, we run both SFT and RL training on top of the GLM-4.5-Air-Base model, scaling RL training up to 512 H200s with high training efficiency.
INTELLECT-2: A Reasoning Model Trained Through Globally Decentralized Reinforcement Learning
May 12, 2025Abstract:We introduce INTELLECT-2, the first globally distributed reinforcement learning (RL) training run of a 32 billion parameter language model. Unlike traditional centralized training efforts, INTELLECT-2 trains a reasoning model using fully asynchronous RL across a dynamic, heterogeneous swarm of permissionless compute contributors. To enable a training run with this unique infrastructure, we built various components from scratch: we introduce PRIME-RL, our training framework purpose-built for distributed asynchronous reinforcement learning, based on top of novel components such as TOPLOC, which verifies rollouts from untrusted inference workers, and SHARDCAST, which efficiently broadcasts policy weights from training nodes to inference workers. Beyond infrastructure components, we propose modifications to the standard GRPO training recipe and data filtering techniques that were crucial to achieve training stability and ensure that our model successfully learned its training objective, thus improving upon QwQ-32B, the state of the art reasoning model in the 32B parameter range. We open-source INTELLECT-2 along with all of our code and data, hoping to encourage and enable more open research in the field of decentralized training.
METAGENE-1: Metagenomic Foundation Model for Pandemic Monitoring
Jan 03, 2025



Abstract:We pretrain METAGENE-1, a 7-billion-parameter autoregressive transformer model, which we refer to as a metagenomic foundation model, on a novel corpus of diverse metagenomic DNA and RNA sequences comprising over 1.5 trillion base pairs. This dataset is sourced from a large collection of human wastewater samples, processed and sequenced using deep metagenomic (next-generation) sequencing methods. Unlike genomic models that focus on individual genomes or curated sets of specific species, the aim of METAGENE-1 is to capture the full distribution of genomic information present within this wastewater, to aid in tasks relevant to pandemic monitoring and pathogen detection. We carry out byte-pair encoding (BPE) tokenization on our dataset, tailored for metagenomic sequences, and then pretrain our model. In this paper, we first detail the pretraining dataset, tokenization strategy, and model architecture, highlighting the considerations and design choices that enable the effective modeling of metagenomic data. We then show results of pretraining this model on our metagenomic dataset, providing details about our losses, system metrics, and training stability over the course of pretraining. Finally, we demonstrate the performance of METAGENE-1, which achieves state-of-the-art results on a set of genomic benchmarks and new evaluations focused on human-pathogen detection and genomic sequence embedding, showcasing its potential for public health applications in pandemic monitoring, biosurveillance, and early detection of emerging health threats.
OpenDiLoCo: An Open-Source Framework for Globally Distributed Low-Communication Training
Jul 10, 2024Abstract:OpenDiLoCo is an open-source implementation and replication of the Distributed Low-Communication (DiLoCo) training method for large language models. We provide a reproducible implementation of the DiLoCo experiments, offering it within a scalable, decentralized training framework using the Hivemind library. We demonstrate its effectiveness by training a model across two continents and three countries, while maintaining 90-95% compute utilization. Additionally, we conduct ablations studies focusing on the algorithm's compute efficiency, scalability in the number of workers and show that its gradients can be all-reduced using FP16 without any performance degradation. Furthermore, we scale OpenDiLoCo to 3x the size of the original work, demonstrating its effectiveness for billion parameter models.
Efficient Parallelization Layouts for Large-Scale Distributed Model Training
Nov 09, 2023Abstract:Efficiently training large language models requires parallelizing across hundreds of hardware accelerators and invoking various compute and memory optimizations. When combined, many of these strategies have complex interactions regarding the final training efficiency. Prior work tackling this problem did not have access to the latest set of optimizations, such as FlashAttention or sequence parallelism. In this work, we conduct a comprehensive ablation study of possible training configurations for large language models. We distill this large study into several key recommendations for the most efficient training. For instance, we find that using a micro-batch size of 1 usually enables the most efficient training layouts. Larger micro-batch sizes necessitate activation checkpointing or higher degrees of model parallelism and also lead to larger pipeline bubbles. Our most efficient configurations enable us to achieve state-of-the-art training efficiency results over a range of model sizes, most notably a Model FLOPs utilization of 70.5% when training a 13B model.
Less Is More: A Comparison of Active Learning Strategies for 3D Medical Image Segmentation
Jul 02, 2022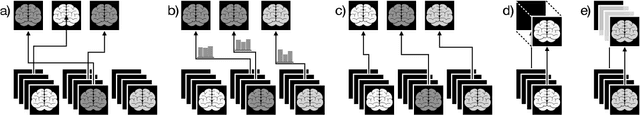
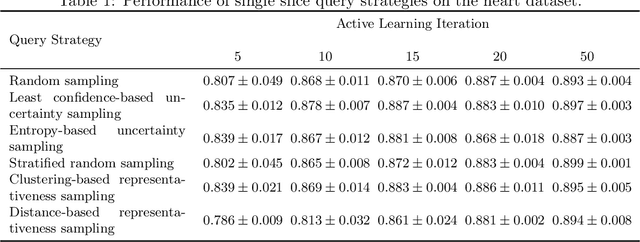
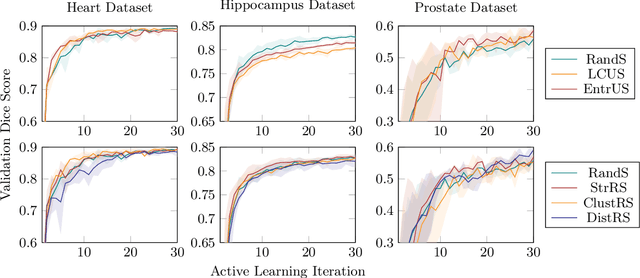
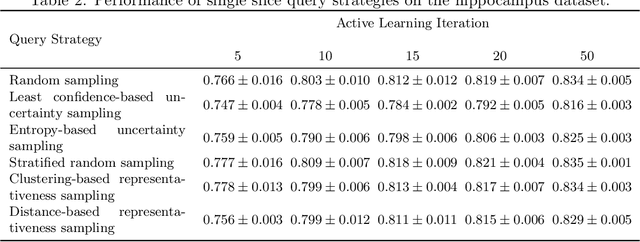
Abstract:Since labeling medical image data is a costly and labor-intensive process, active learning has gained much popularity in the medical image segmentation domain in recent years. A variety of active learning strategies have been proposed in the literature, but their effectiveness is highly dependent on the dataset and training scenario. To facilitate the comparison of existing strategies and provide a baseline for evaluating novel strategies, we evaluate the performance of several well-known active learning strategies on three datasets from the Medical Segmentation Decathlon. Additionally, we consider a strided sampling strategy specifically tailored to 3D image data. We demonstrate that both random and strided sampling act as strong baselines and discuss the advantages and disadvantages of the studied methods. To allow other researchers to compare their work to our results, we provide an open-source framework for benchmarking active learning strategies on a variety of medical segmentation datasets.
Parallel Statistical Multi-resolution Estimation
Mar 10, 2015



Abstract:We discuss several strategies to implement Dykstra's projection algorithm on NVIDIA's compute unified device architecture (CUDA). Dykstra's algorithm is the central step in and the computationally most expensive part of statistical multi-resolution methods. It projects a given vector onto the intersection of convex sets. Compared with a CPU implementation our CUDA implementation is one order of magnitude faster. For a further speed up and to reduce memory consumption we have developed a new variant, which we call incomplete Dykstra's algorithm. Implemented in CUDA it is one order of magnitude faster than the CUDA implementation of the standard Dykstra algorithm. As sample application we discuss using the incomplete Dykstra's algorithm as preprocessor for the recently developed super-resolution optical fluctuation imaging (SOFI) method (Dertinger et al. 2009). We show that statistical multi-resolution estimation can enhance the resolution improvement of the plain SOFI algorithm just as the Fourier-reweighting of SOFI. The results are compared in terms of their power spectrum and their Fourier ring correlation (Saxton and Baumeister 1982). The Fourier ring correlation indicates that the resolution for typical second order SOFI images can be improved by about 30 per cent. Our results show that a careful parallelization of Dykstra's algorithm enables its use in large-scale statistical multi-resolution analyses.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge