Jintao Meng
Taylor-Sensus Network: Embracing Noise to Enlighten Uncertainty for Scientific Data
Sep 12, 2024



Abstract:Uncertainty estimation is crucial in scientific data for machine learning. Current uncertainty estimation methods mainly focus on the model's inherent uncertainty, while neglecting the explicit modeling of noise in the data. Furthermore, noise estimation methods typically rely on temporal or spatial dependencies, which can pose a significant challenge in structured scientific data where such dependencies among samples are often absent. To address these challenges in scientific research, we propose the Taylor-Sensus Network (TSNet). TSNet innovatively uses a Taylor series expansion to model complex, heteroscedastic noise and proposes a deep Taylor block for aware noise distribution. TSNet includes a noise-aware contrastive learning module and a data density perception module for aleatoric and epistemic uncertainty. Additionally, an uncertainty combination operator is used to integrate these uncertainties, and the network is trained using a novel heteroscedastic mean square error loss. TSNet demonstrates superior performance over mainstream and state-of-the-art methods in experiments, highlighting its potential in scientific research and noise resistance. It will be open-source to facilitate the community of "AI for Science".
Autism Spectrum Disorder Classification in Children based on Structural MRI Features Extracted using Contrastive Variational Autoencoder
Jul 03, 2023
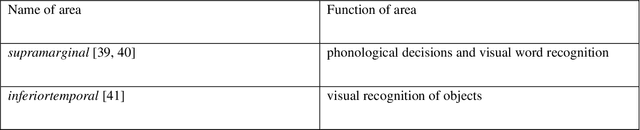


Abstract:Autism spectrum disorder (ASD) is a highly disabling mental disease that brings significant impairments of social interaction ability to the patients, making early screening and intervention of ASD critical. With the development of the machine learning and neuroimaging technology, extensive research has been conducted on machine classification of ASD based on structural MRI (s-MRI). However, most studies involve with datasets where participants' age are above 5. Few studies conduct machine classification of ASD for participants below 5-year-old, but, with mediocre predictive accuracy. In this paper, we push the boundary of predictive accuracy (above 0.97) of machine classification of ASD in children (age range: 0.92-4.83 years), based on s-MRI features extracted using contrastive variational autoencoder (CVAE). 78 s-MRI, collected from Shenzhen Children's Hospital, are used for training CVAE, which consists of both ASD-specific feature channel and common shared feature channel. The ASD participants represented by ASD-specific features can be easily discriminated from TC participants represented by the common shared features, leading to high classification accuracy. In case of degraded predictive accuracy when data size is extremely small, a transfer learning strategy is proposed here as a potential solution. Finally, we conduct neuroanatomical interpretation based on the correlation between s-MRI features extracted from CVAE and surface area of different cortical regions, which discloses potential biomarkers that could help target treatments of ASD in the future.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge