Jing Xing
Boosting Single-domain Generalized Object Detection via Vision-Language Knowledge Interaction
Apr 27, 2025
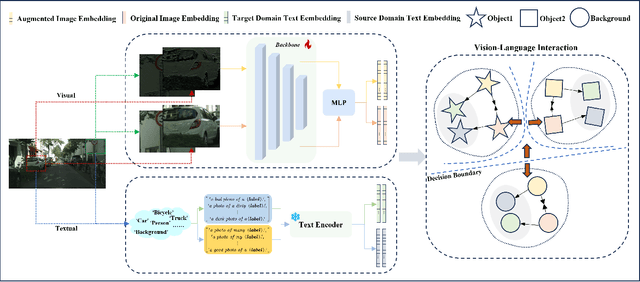


Abstract:Single-Domain Generalized Object Detection~(S-DGOD) aims to train an object detector on a single source domain while generalizing well to diverse unseen target domains, making it suitable for multimedia applications that involve various domain shifts, such as intelligent video surveillance and VR/AR technologies. With the success of large-scale Vision-Language Models, recent S-DGOD approaches exploit pre-trained vision-language knowledge to guide invariant feature learning across visual domains. However, the utilized knowledge remains at a coarse-grained level~(e.g., the textual description of adverse weather paired with the image) and serves as an implicit regularization for guidance, struggling to learn accurate region- and object-level features in varying domains. In this work, we propose a new cross-modal feature learning method, which can capture generalized and discriminative regional features for S-DGOD tasks. The core of our method is the mechanism of Cross-modal and Region-aware Feature Interaction, which simultaneously learns both inter-modal and intra-modal regional invariance through dynamic interactions between fine-grained textual and visual features. Moreover, we design a simple but effective strategy called Cross-domain Proposal Refining and Mixing, which aligns the position of region proposals across multiple domains and diversifies them, enhancing the localization ability of detectors in unseen scenarios. Our method achieves new state-of-the-art results on S-DGOD benchmark datasets, with improvements of +8.8\%~mPC on Cityscapes-C and +7.9\%~mPC on DWD over baselines, demonstrating its efficacy.
MoCL: Contrastive Learning on Molecular Graphs with Multi-level Domain Knowledge
Jun 05, 2021



Abstract:Recent years have seen a rapid growth of utilizing graph neural networks (GNNs) in the biomedical domain for tackling drug-related problems. However, like any other deep architectures, GNNs are data hungry. While requiring labels in real world is often expensive, pretraining GNNs in an unsupervised manner has been actively explored. Among them, graph contrastive learning, by maximizing the mutual information between paired graph augmentations, has been shown to be effective on various downstream tasks. However, the current graph contrastive learning framework has two limitations. First, the augmentations are designed for general graphs and thus may not be suitable or powerful enough for certain domains. Second, the contrastive scheme only learns representations that are invariant to local perturbations and thus does not consider the global structure of the dataset, which may also be useful for downstream tasks. Therefore, in this paper, we study graph contrastive learning in the context of biomedical domain, where molecular graphs are present. We propose a novel framework called MoCL, which utilizes domain knowledge at both local- and global-level to assist representation learning. The local-level domain knowledge guides the augmentation process such that variation is introduced without changing graph semantics. The global-level knowledge encodes the similarity information between graphs in the entire dataset and helps to learn representations with richer semantics. The entire model is learned through a double contrast objective. We evaluate MoCL on various molecular datasets under both linear and semi-supervised settings and results show that MoCL achieves state-of-the-art performance.
Robust Collaborative Learning with Noisy Labels
Dec 26, 2020



Abstract:Learning with curriculum has shown great effectiveness in tasks where the data contains noisy (corrupted) labels, since the curriculum can be used to re-weight or filter out noisy samples via proper design. However, obtaining curriculum from a learner itself without additional supervision or feedback deteriorates the effectiveness due to sample selection bias. Therefore, methods that involve two or more networks have been recently proposed to mitigate such bias. Nevertheless, these studies utilize the collaboration between networks in a way that either emphasizes the disagreement or focuses on the agreement while ignores the other. In this paper, we study the underlying mechanism of how disagreement and agreement between networks can help reduce the noise in gradients and develop a novel framework called Robust Collaborative Learning (RCL) that leverages both disagreement and agreement among networks. We demonstrate the effectiveness of RCL on both synthetic benchmark image data and real-world large-scale bioinformatics data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge