Jiayang Gao
HaluNet: Multi-Granular Uncertainty Modeling for Efficient Hallucination Detection in LLM Question Answering
Dec 31, 2025Abstract:Large Language Models (LLMs) excel at question answering (QA) but often generate hallucinations, including factual errors or fabricated content. Detecting hallucinations from internal uncertainty signals is attractive due to its scalability and independence from external resources. Existing methods often aim to accurately capture a single type of uncertainty while overlooking the complementarity among different sources, particularly between token-level probability uncertainty and the uncertainty conveyed by internal semantic representations, which provide complementary views on model reliability. We present \textbf{HaluNet}, a lightweight and trainable neural framework that integrates multi granular token level uncertainties by combining semantic embeddings with probabilistic confidence and distributional uncertainty. Its multi branch architecture adaptively fuses what the model knows with the uncertainty expressed in its outputs, enabling efficient one pass hallucination detection. Experiments on SQuAD, TriviaQA, and Natural Questions show that HaluNet delivers strong detection performance and favorable computational efficiency, with or without access to context, highlighting its potential for real time hallucination detection in LLM based QA systems.
Where We Have Arrived in Proving the Emergence of Sparse Symbolic Concepts in AI Models
May 03, 2023

Abstract:This paper aims to prove the emergence of symbolic concepts in well-trained AI models. We prove that if (1) the high-order derivatives of the model output w.r.t. the input variables are all zero, (2) the AI model can be used on occluded samples and will yield higher confidence when the input sample is less occluded, and (3) the confidence of the AI model does not significantly degrade on occluded samples, then the AI model will encode sparse interactive concepts. Each interactive concept represents an interaction between a specific set of input variables, and has a certain numerical effect on the inference score of the model. Specifically, it is proved that the inference score of the model can always be represented as the sum of the interaction effects of all interactive concepts. In fact, we hope to prove that conditions for the emergence of symbolic concepts are quite common. It means that for most AI models, we can usually use a small number of interactive concepts to mimic the model outputs on any arbitrarily masked samples.
Predictive models of RNA degradation through dual crowdsourcing
Oct 14, 2021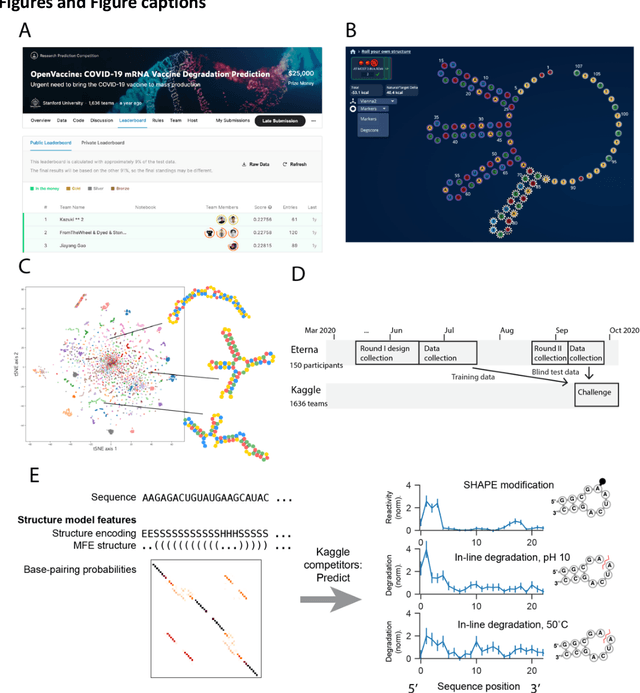
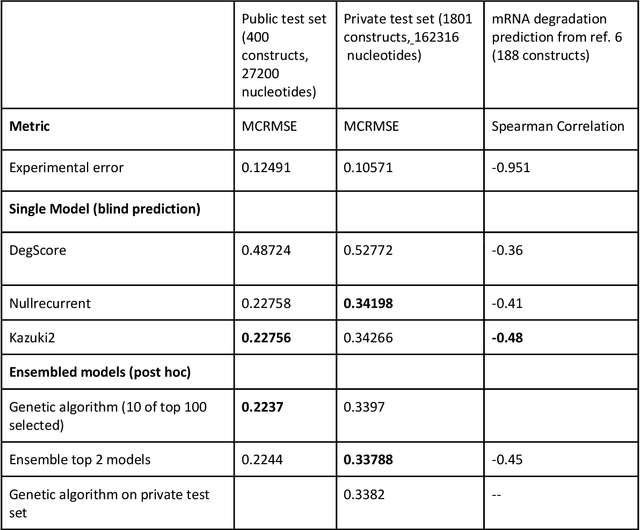
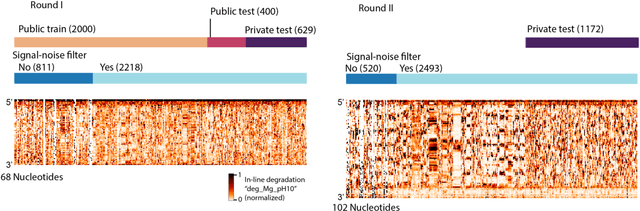
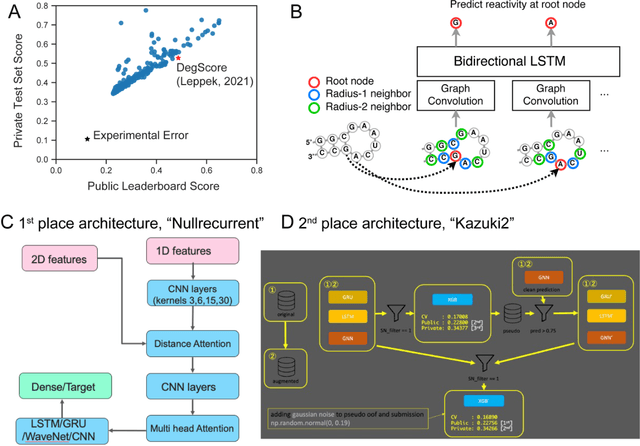
Abstract:Messenger RNA-based medicines hold immense potential, as evidenced by their rapid deployment as COVID-19 vaccines. However, worldwide distribution of mRNA molecules has been limited by their thermostability, which is fundamentally limited by the intrinsic instability of RNA molecules to a chemical degradation reaction called in-line hydrolysis. Predicting the degradation of an RNA molecule is a key task in designing more stable RNA-based therapeutics. Here, we describe a crowdsourced machine learning competition ("Stanford OpenVaccine") on Kaggle, involving single-nucleotide resolution measurements on 6043 102-130-nucleotide diverse RNA constructs that were themselves solicited through crowdsourcing on the RNA design platform Eterna. The entire experiment was completed in less than 6 months. Winning models demonstrated test set errors that were better by 50% than the previous state-of-the-art DegScore model. Furthermore, these models generalized to blindly predicting orthogonal degradation data on much longer mRNA molecules (504-1588 nucleotides) with improved accuracy over DegScore and other models. Top teams integrated natural language processing architectures and data augmentation techniques with predictions from previous dynamic programming models for RNA secondary structure. These results indicate that such models are capable of representing in-line hydrolysis with excellent accuracy, supporting their use for designing stabilized messenger RNAs. The integration of two crowdsourcing platforms, one for data set creation and another for machine learning, may be fruitful for other urgent problems that demand scientific discovery on rapid timescales.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge