Jiashuang Huang
M2EF-NNs: Multimodal Multi-instance Evidence Fusion Neural Networks for Cancer Survival Prediction
Aug 08, 2024Abstract:Accurate cancer survival prediction is crucial for assisting clinical doctors in formulating treatment plans. Multimodal data, including histopathological images and genomic data, offer complementary and comprehensive information that can greatly enhance the accuracy of this task. However, the current methods, despite yielding promising results, suffer from two notable limitations: they do not effectively utilize global context and disregard modal uncertainty. In this study, we put forward a neural network model called M2EF-NNs, which leverages multimodal and multi-instance evidence fusion techniques for accurate cancer survival prediction. Specifically, to capture global information in the images, we use a pre-trained Vision Transformer (ViT) model to obtain patch feature embeddings of histopathological images. Then, we introduce a multimodal attention module that uses genomic embeddings as queries and learns the co-attention mapping between genomic and histopathological images to achieve an early interaction fusion of multimodal information and better capture their correlations. Subsequently, we are the first to apply the Dempster-Shafer evidence theory (DST) to cancer survival prediction. We parameterize the distribution of class probabilities using the processed multimodal features and introduce subjective logic to estimate the uncertainty associated with different modalities. By combining with the Dempster-Shafer theory, we can dynamically adjust the weights of class probabilities after multimodal fusion to achieve trusted survival prediction. Finally, Experimental validation on the TCGA datasets confirms the significant improvements achieved by our proposed method in cancer survival prediction and enhances the reliability of the model.
FDiff-Fusion:Denoising diffusion fusion network based on fuzzy learning for 3D medical image segmentation
Jul 22, 2024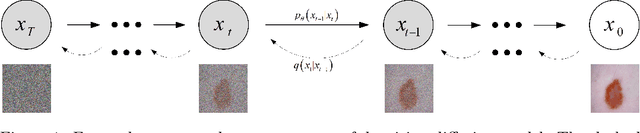
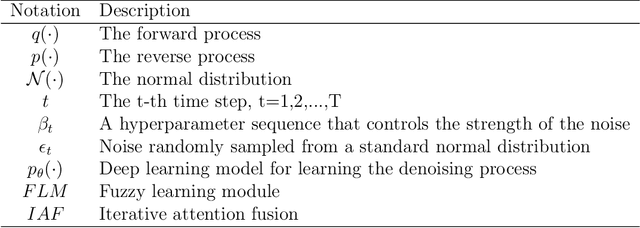
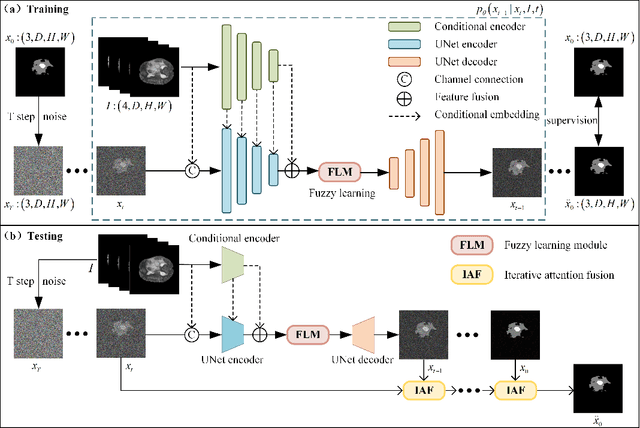
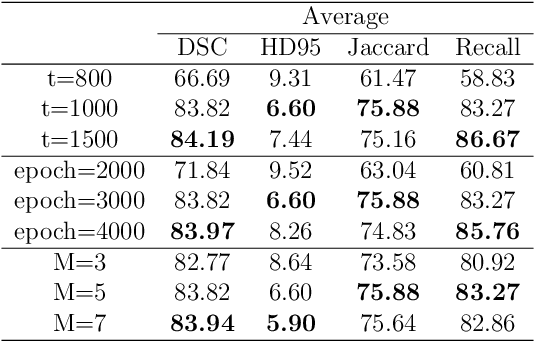
Abstract:In recent years, the denoising diffusion model has achieved remarkable success in image segmentation modeling. With its powerful nonlinear modeling capabilities and superior generalization performance, denoising diffusion models have gradually been applied to medical image segmentation tasks, bringing new perspectives and methods to this field. However, existing methods overlook the uncertainty of segmentation boundaries and the fuzziness of regions, resulting in the instability and inaccuracy of the segmentation results. To solve this problem, a denoising diffusion fusion network based on fuzzy learning for 3D medical image segmentation (FDiff-Fusion) is proposed in this paper. By integrating the denoising diffusion model into the classical U-Net network, this model can effectively extract rich semantic information from input medical images, thus providing excellent pixel-level representation for medical image segmentation. ... Finally, to validate the effectiveness of FDiff-Fusion, we compare it with existing advanced segmentation networks on the BRATS 2020 brain tumor dataset and the BTCV abdominal multi-organ dataset. The results show that FDiff-Fusion significantly improves the Dice scores and HD95 distance on these two datasets, demonstrating its superiority in medical image segmentation tasks.
* This paper has been accepted by Information Fusion. Permission from Elsevier must be obtained for all other uses, in any current or future media. The final version is available at [doi:10.1016/J.INFFUS.2024.102540]
Cascaded two-stage feature clustering and selection via separability and consistency in fuzzy decision systems
Jul 22, 2024



Abstract:Feature selection is a vital technique in machine learning, as it can reduce computational complexity, improve model performance, and mitigate the risk of overfitting. However, the increasing complexity and dimensionality of datasets pose significant challenges in the selection of features. Focusing on these challenges, this paper proposes a cascaded two-stage feature clustering and selection algorithm for fuzzy decision systems. In the first stage, we reduce the search space by clustering relevant features and addressing inter-feature redundancy. In the second stage, a clustering-based sequentially forward selection method that explores the global and local structure of data is presented. We propose a novel metric for assessing the significance of features, which considers both global separability and local consistency. Global separability measures the degree of intra-class cohesion and inter-class separation based on fuzzy membership, providing a comprehensive understanding of data separability. Meanwhile, local consistency leverages the fuzzy neighborhood rough set model to capture uncertainty and fuzziness in the data. The effectiveness of our proposed algorithm is evaluated through experiments conducted on 18 public datasets and a real-world schizophrenia dataset. The experiment results demonstrate our algorithm's superiority over benchmarking algorithms in both classification accuracy and the number of selected features.
* This paper has been accepted by IEEE Transactions on Fuzzy Systems for publication. Permission from IEEE must be obtained for all other uses, in any current or future media. The final version is available at [10.1109/TFUZZ.2024.3420963]
FMDNN: A Fuzzy-guided Multi-granular Deep Neural Network for Histopathological Image Classification
Jul 22, 2024



Abstract:Histopathological image classification constitutes a pivotal task in computer-aided diagnostics. The precise identification and categorization of histopathological images are of paramount significance for early disease detection and treatment. In the diagnostic process of pathologists, a multi-tiered approach is typically employed to assess abnormalities in cell regions at different magnifications. However, feature extraction is often performed at a single granularity, overlooking the multi-granular characteristics of cells. To address this issue, we propose the Fuzzy-guided Multi-granularity Deep Neural Network (FMDNN). Inspired by the multi-granular diagnostic approach of pathologists, we perform feature extraction on cell structures at coarse, medium, and fine granularity, enabling the model to fully harness the information in histopathological images. We incorporate the theory of fuzzy logic to address the challenge of redundant key information arising during multi-granular feature extraction. Cell features are described from different perspectives using multiple fuzzy membership functions, which are fused to create universal fuzzy features. A fuzzy-guided cross-attention module guides universal fuzzy features toward multi-granular features. We propagate these features through an encoder to all patch tokens, aiming to achieve enhanced classification accuracy and robustness. In experiments on multiple public datasets, our model exhibits a significant improvement in accuracy over commonly used classification methods for histopathological image classification and shows commendable interpretability.
* This paper has been accepted by IEEE Transactions on Fuzzy Systems for publication. Permission from IEEE must be obtained for all other uses, in any current or future media. The final version is available at [doi: 10.1109/TFUZZ.2024.3410929]
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge