Jialin Peng
BiPrompt-SAM: Enhancing Image Segmentation via Explicit Selection between Point and Text Prompts
Mar 25, 2025



Abstract:Segmentation is a fundamental task in computer vision, with prompt-driven methods gaining prominence due to their flexibility. The recent Segment Anything Model (SAM) has demonstrated powerful point-prompt segmentation capabilities, while text-based segmentation models offer rich semantic understanding. However, existing approaches rarely explore how to effectively combine these complementary modalities for optimal segmentation performance. This paper presents BiPrompt-SAM, a novel dual-modal prompt segmentation framework that fuses the advantages of point and text prompts through an explicit selection mechanism. Specifically, we leverage SAM's inherent ability to generate multiple mask candidates, combined with a semantic guidance mask from text prompts, and explicitly select the most suitable candidate based on similarity metrics. This approach can be viewed as a simplified Mixture of Experts (MoE) system, where the point and text modules act as distinct "experts," and the similarity scoring serves as a rudimentary "gating network." We conducted extensive evaluations on both the Endovis17 medical dataset and RefCOCO series natural image datasets. On Endovis17, BiPrompt-SAM achieved 89.55\% mDice and 81.46\% mIoU, comparable to state-of-the-art specialized medical segmentation models. On the RefCOCO series datasets, our method attained 87.1\%, 86.5\%, and 85.8\% IoU, significantly outperforming existing approaches. Experiments demonstrate that our explicit dual-selection method effectively combines the spatial precision of point prompts with the semantic richness of text prompts, particularly excelling in scenarios involving semantically complex objects, multiple similar objects, and partial occlusions. BiPrompt-SAM not only provides a simple yet effective implementation but also offers a new perspective on multi-modal prompt fusion.
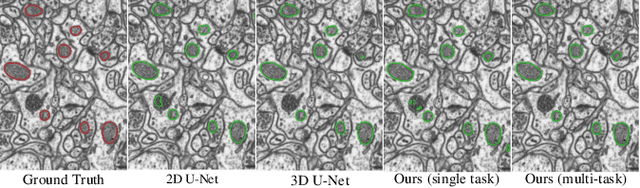
Weakly-Supervised Cross-Domain Segmentation of Electron Microscopy with Sparse Point Annotation
Mar 31, 2024



Abstract:Accurate segmentation of organelle instances from electron microscopy (EM) images plays an essential role in many neuroscience researches. However, practical scenarios usually suffer from high annotation costs, label scarcity, and large domain diversity. While unsupervised domain adaptation (UDA) that assumes no annotation effort on the target data is promising to alleviate these challenges, its performance on complicated segmentation tasks is still far from practical usage. To address these issues, we investigate a highly annotation-efficient weak supervision, which assumes only sparse center-points on a small subset of object instances in the target training images. To achieve accurate segmentation with partial point annotations, we introduce instance counting and center detection as auxiliary tasks and design a multitask learning framework to leverage correlations among the counting, detection, and segmentation, which are all tasks with partial or no supervision. Building upon the different domain-invariances of the three tasks, we enforce counting estimation with a novel soft consistency loss as a global prior for center detection, which further guides the per-pixel segmentation. To further compensate for annotation sparsity, we develop a cross-position cut-and-paste for label augmentation and an entropy-based pseudo-label selection. The experimental results highlight that, by simply using extremely weak annotation, e.g., 15\% sparse points, for model training, the proposed model is capable of significantly outperforming UDA methods and produces comparable performance as the supervised counterpart. The high robustness of our model shown in the validations and the low requirement of expert knowledge for sparse point annotation further improve the potential application value of our model.
WDA-Net: Weakly-Supervised Domain Adaptive Segmentation of Electron Microscopy
Oct 30, 2022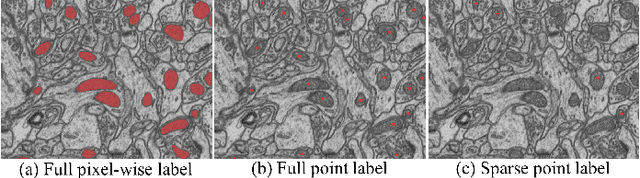
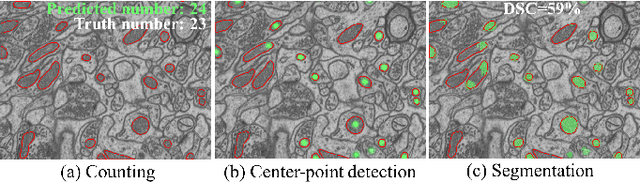
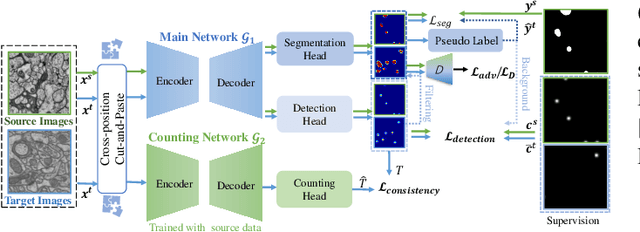
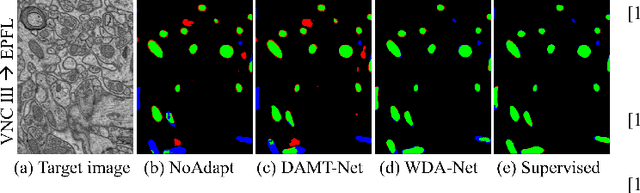
Abstract:Accurate segmentation of organelle instances, e.g., mitochondria, is essential for electron microscopy analysis. Despite the outstanding performance of fully supervised methods, they highly rely on sufficient per-pixel annotated data and are sensitive to domain shift. Aiming to develop a highly annotation-efficient approach with competitive performance, we focus on weakly-supervised domain adaptation (WDA) with a type of extremely sparse and weak annotation demanding minimal annotation efforts, i.e., sparse point annotations on only a small subset of object instances. To reduce performance degradation arising from domain shift, we explore multi-level transferable knowledge through conducting three complementary tasks, i.e., counting, detection, and segmentation, constituting a task pyramid with different levels of domain invariance. The intuition behind this is that after investigating a related source domain, it is much easier to spot similar objects in the target domain than to delineate their fine boundaries. Specifically, we enforce counting estimation as a global constraint to the detection with sparse supervision, which further guides the segmentation. A cross-position cut-and-paste augmentation is introduced to further compensate for the annotation sparsity. Extensive validations show that our model with only 15% point annotations can achieve comparable performance as supervised models and shows robustness to annotation selection.
Medical Image Segmentation with Limited Supervision: A Review of Deep Network Models
Feb 28, 2021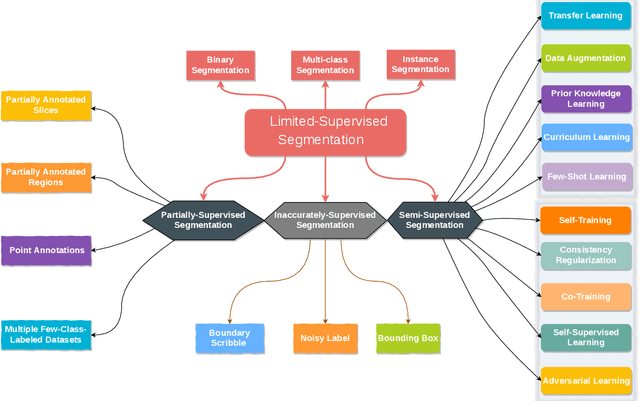
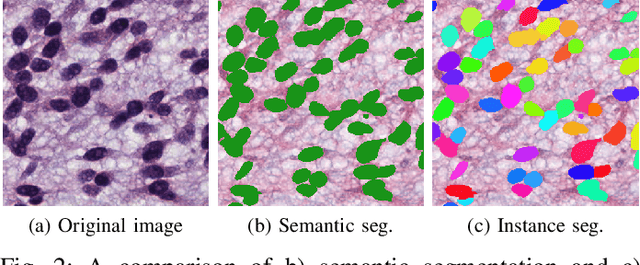
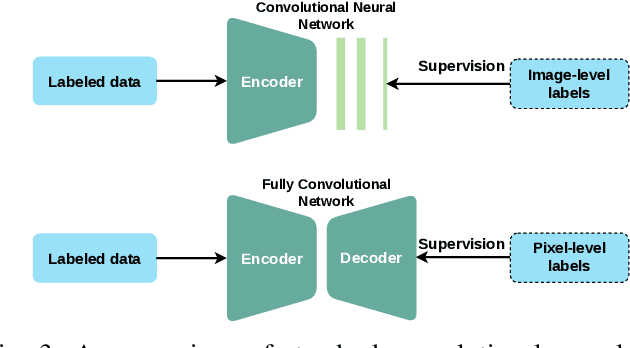
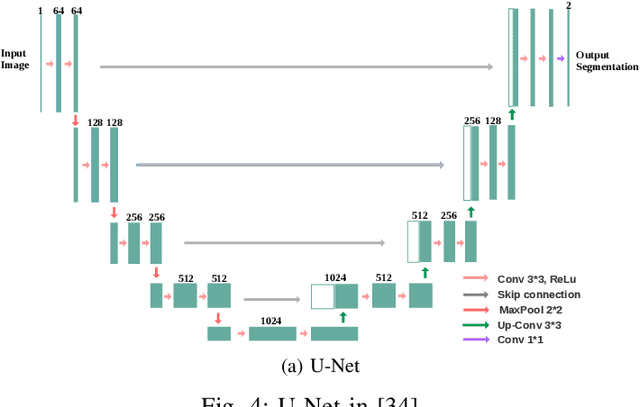
Abstract:Despite the remarkable performance of deep learning methods on various tasks, most cutting-edge models rely heavily on large-scale annotated training examples, which are often unavailable for clinical and health care tasks. The labeling costs for medical images are very high, especially in medical image segmentation, which typically requires intensive pixel/voxel-wise labeling. Therefore, the strong capability of learning and generalizing from limited supervision, including a limited amount of annotations, sparse annotations, and inaccurate annotations, is crucial for the successful application of deep learning models in medical image segmentation. However, due to its intrinsic difficulty, segmentation with limited supervision is challenging and specific model design and/or learning strategies are needed. In this paper, we provide a systematic and up-to-date review of the solutions above, with summaries and comments about the methodologies. We also highlight several problems in this field, discussed future directions observing further investigations.
HIVE-Net: Centerline-Aware HIerarchical View-Ensemble Convolutional Network for Mitochondria Segmentation in EM Images
Jan 08, 2021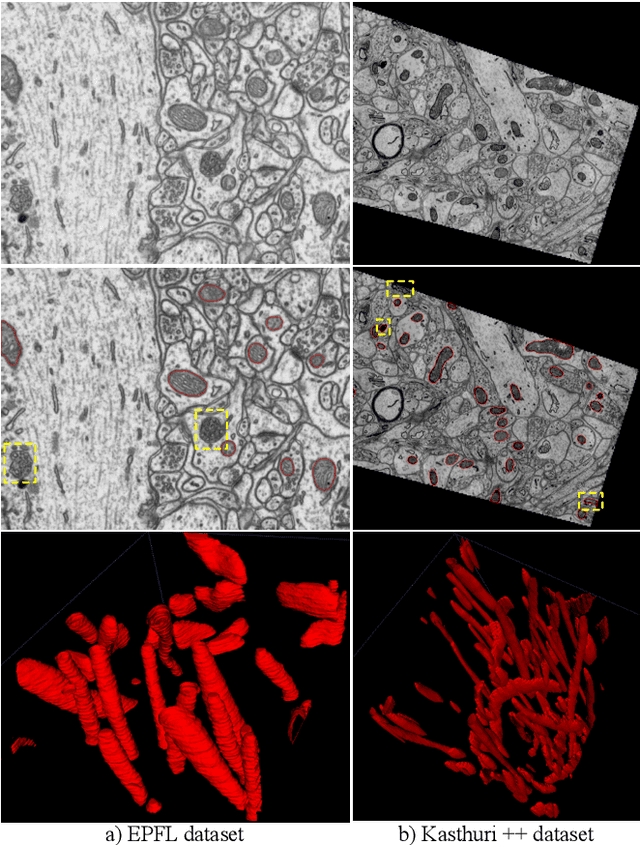
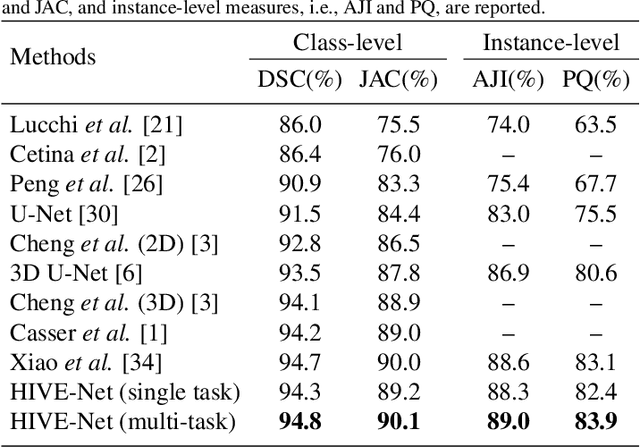
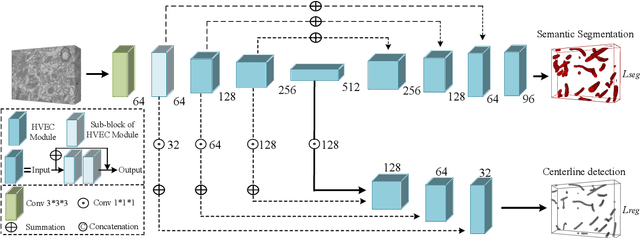

Abstract:Semantic segmentation of electron microscopy (EM) is an essential step to efficiently obtain reliable morphological statistics. Despite the great success achieved using deep convolutional neural networks (CNNs), they still produce coarse segmentations with lots of discontinuities and false positives for mitochondria segmentation. In this study, we introduce a centerline-aware multitask network by utilizing centerline as an intrinsic shape cue of mitochondria to regularize the segmentation. Since the application of 3D CNNs on large medical volumes is usually hindered by their substantial computational cost and storage overhead, we introduce a novel hierarchical view-ensemble convolution (HVEC), a simple alternative of 3D convolution to learn 3D spatial contexts using more efficient 2D convolutions. The HVEC enables both decomposing and sharing multi-view information, leading to increased learning capacity. Extensive validation results on two challenging benchmarks show that, the proposed method performs favorably against the state-of-the-art methods in accuracy and visual quality but with a greatly reduced model size. Moreover, the proposed model also shows significantly improved generalization ability, especially when training with quite limited amount of training data.
Adversarial-Prediction Guided Multi-task Adaptation for Semantic Segmentation of Electron Microscopy Images
Apr 05, 2020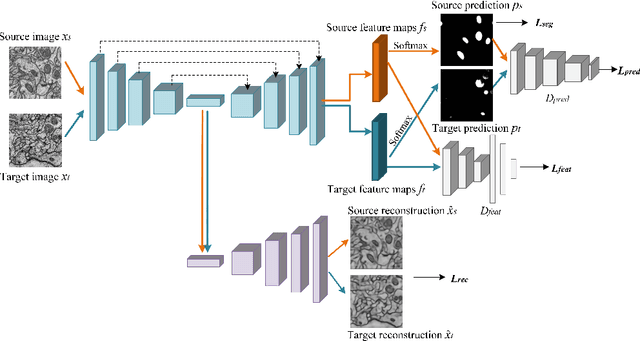
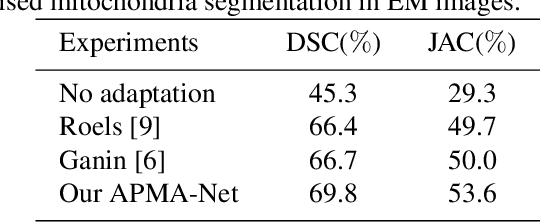


Abstract:Semantic segmentation is an essential step for electron microscopy (EM) image analysis. Although supervised models have achieved significant progress, the need for labor intensive pixel-wise annotation is a major limitation. To complicate matters further, supervised learning models may not generalize well on a novel dataset due to domain shift. In this study, we introduce an adversarial-prediction guided multi-task network to learn the adaptation of a well-trained model for use on a novel unlabeled target domain. Since no label is available on target domain, we learn an encoding representation not only for the supervised segmentation on source domain but also for unsupervised reconstruction of the target data. To improve the discriminative ability with geometrical cues, we further guide the representation learning by multi-level adversarial learning in semantic prediction space. Comparisons and ablation study on public benchmark demonstrated state-of-the-art performance and effectiveness of our approach.
EM-NET: Centerline-Aware Mitochondria Segmentation in EM Images via Hierarchical View-Ensemble Convolutional Network
Jan 09, 2020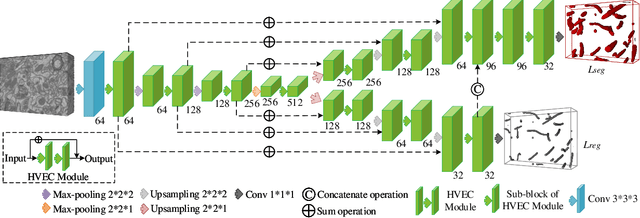
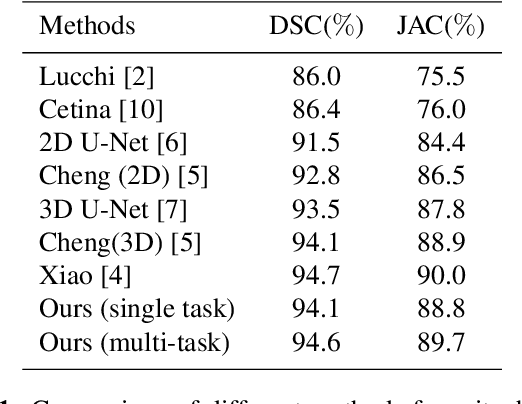
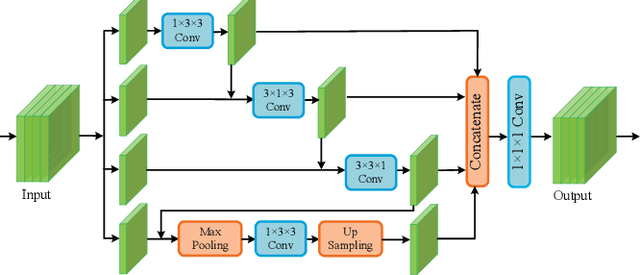
Abstract:Although deep encoder-decoder networks have achieved astonishing performance for mitochondria segmentation from electron microscopy (EM) images, they still produce coarse segmentations with lots of discontinuities and false positives. Besides, the need for labor intensive annotations of large 3D dataset and huge memory overhead by 3D models are also major limitations. To address these problems, we introduce a multi-task network named EM-Net, which includes an auxiliary centerline detection task to account for shape information of mitochondria represented by centerline. Therefore, the centerline detection sub-network is able to enhance the accuracy and robustness of segmentation task, especially when only a small set of annotated data are available. To achieve a light-weight 3D network, we introduce a novel hierarchical view-ensemble convolution module to reduce number of parameters, and facilitate multi-view information aggregation.Validations on public benchmark showed state-of-the-art performance by EM-Net. Even with significantly reduced training data, our method still showed quite promising results.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge