WDA-Net: Weakly-Supervised Domain Adaptive Segmentation of Electron Microscopy
Paper and Code
Oct 30, 2022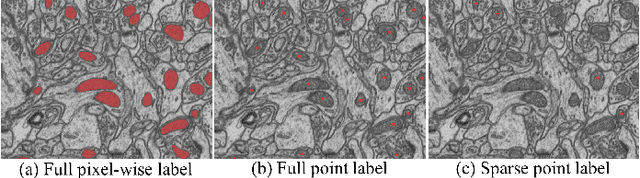
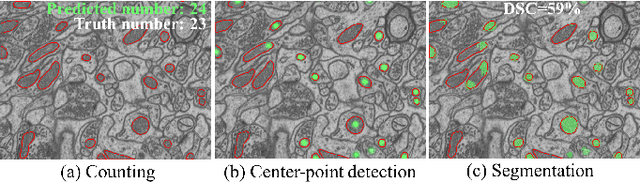
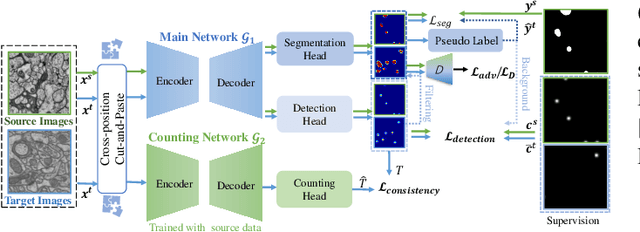
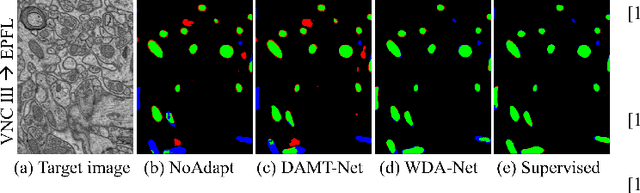
Accurate segmentation of organelle instances, e.g., mitochondria, is essential for electron microscopy analysis. Despite the outstanding performance of fully supervised methods, they highly rely on sufficient per-pixel annotated data and are sensitive to domain shift. Aiming to develop a highly annotation-efficient approach with competitive performance, we focus on weakly-supervised domain adaptation (WDA) with a type of extremely sparse and weak annotation demanding minimal annotation efforts, i.e., sparse point annotations on only a small subset of object instances. To reduce performance degradation arising from domain shift, we explore multi-level transferable knowledge through conducting three complementary tasks, i.e., counting, detection, and segmentation, constituting a task pyramid with different levels of domain invariance. The intuition behind this is that after investigating a related source domain, it is much easier to spot similar objects in the target domain than to delineate their fine boundaries. Specifically, we enforce counting estimation as a global constraint to the detection with sparse supervision, which further guides the segmentation. A cross-position cut-and-paste augmentation is introduced to further compensate for the annotation sparsity. Extensive validations show that our model with only 15% point annotations can achieve comparable performance as supervised models and shows robustness to annotation selection.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge