Jiakang Chen
TTT-Parkour: Rapid Test-Time Training for Perceptive Robot Parkour
Feb 02, 2026Abstract:Achieving highly dynamic humanoid parkour on unseen, complex terrains remains a challenge in robotics. Although general locomotion policies demonstrate capabilities across broad terrain distributions, they often struggle with arbitrary and highly challenging environments. To overcome this limitation, we propose a real-to-sim-to-real framework that leverages rapid test-time training (TTT) on novel terrains, significantly enhancing the robot's capability to traverse extremely difficult geometries. We adopt a two-stage end-to-end learning paradigm: a policy is first pre-trained on diverse procedurally generated terrains, followed by rapid fine-tuning on high-fidelity meshes reconstructed from real-world captures. Specifically, we develop a feed-forward, efficient, and high-fidelity geometry reconstruction pipeline using RGB-D inputs, ensuring both speed and quality during test-time training. We demonstrate that TTT-Parkour empowers humanoid robots to master complex obstacles, including wedges, stakes, boxes, trapezoids, and narrow beams. The whole pipeline of capturing, reconstructing, and test-time training requires less than 10 minutes on most tested terrains. Extensive experiments show that the policy after test-time training exhibits robust zero-shot sim-to-real transfer capability.
MEAN-RIR: Multi-Modal Environment-Aware Network for Robust Room Impulse Response Estimation
Sep 05, 2025Abstract:This paper presents a Multi-Modal Environment-Aware Network (MEAN-RIR), which uses an encoder-decoder framework to predict room impulse response (RIR) based on multi-level environmental information from audio, visual, and textual sources. Specifically, reverberant speech capturing room acoustic properties serves as the primary input, which is combined with panoramic images and text descriptions as supplementary inputs. Each input is processed by its respective encoder, and the outputs are fed into cross-attention modules to enable effective interaction between different modalities. The MEAN-RIR decoder generates two distinct components: the first component captures the direct sound and early reflections, while the second produces masks that modulate learnable filtered noise to synthesize the late reverberation. These two components are mixed to reconstruct the final RIR. The results show that MEAN-RIR significantly improves RIR estimation, with notable gains in acoustic parameters.
SING: Semantic Image Communications using Null-Space and INN-Guided Diffusion Models
Mar 16, 2025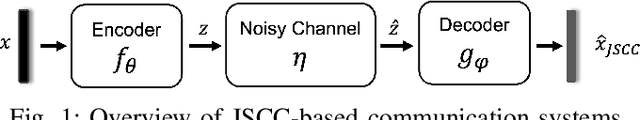
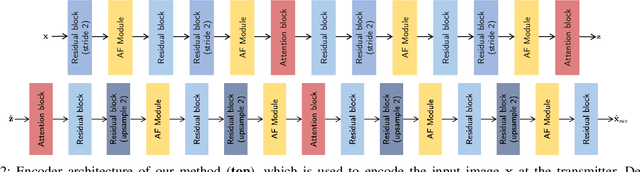
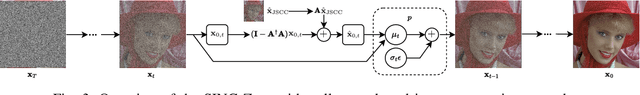
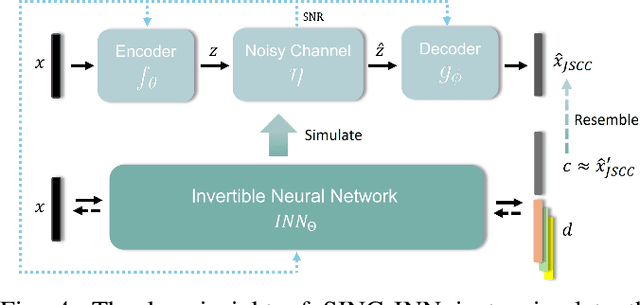
Abstract:Joint source-channel coding systems based on deep neural networks (DeepJSCC) have recently demonstrated remarkable performance in wireless image transmission. Existing methods primarily focus on minimizing distortion between the transmitted image and the reconstructed version at the receiver, often overlooking perceptual quality. This can lead to severe perceptual degradation when transmitting images under extreme conditions, such as low bandwidth compression ratios (BCRs) and low signal-to-noise ratios (SNRs). In this work, we propose SING, a novel two-stage JSCC framework that formulates the recovery of high-quality source images from corrupted reconstructions as an inverse problem. Depending on the availability of information about the DeepJSCC encoder/decoder and the channel at the receiver, SING can either approximate the stochastic degradation as a linear transformation, or leverage invertible neural networks (INNs) for precise modeling. Both approaches enable the seamless integration of diffusion models into the reconstruction process, enhancing perceptual quality. Experimental results demonstrate that SING outperforms DeepJSCC and other approaches, delivering superior perceptual quality even under extremely challenging conditions, including scenarios with significant distribution mismatches between the training and test data.
CommIN: Semantic Image Communications as an Inverse Problem with INN-Guided Diffusion Models
Oct 02, 2023Abstract:Joint source-channel coding schemes based on deep neural networks (DeepJSCC) have recently achieved remarkable performance for wireless image transmission. However, these methods usually focus only on the distortion of the reconstructed signal at the receiver side with respect to the source at the transmitter side, rather than the perceptual quality of the reconstruction which carries more semantic information. As a result, severe perceptual distortion can be introduced under extreme conditions such as low bandwidth and low signal-to-noise ratio. In this work, we propose CommIN, which views the recovery of high-quality source images from degraded reconstructions as an inverse problem. To address this, CommIN combines Invertible Neural Networks (INN) with diffusion models, aiming for superior perceptual quality. Through experiments, we show that our CommIN significantly improves the perceptual quality compared to DeepJSCC under extreme conditions and outperforms other inverse problem approaches used in DeepJSCC.
BioCoder: A Benchmark for Bioinformatics Code Generation with Contextual Pragmatic Knowledge
Sep 05, 2023Abstract:Pre-trained language models like ChatGPT have significantly improved code generation. As these models scale up, there is an increasing need for the output to handle more intricate tasks. Moreover, in bioinformatics, generating functional programs poses additional notable challenges due to the amount of domain knowledge, the need for complicated data operations, and intricate functional dependencies between the operations. Here, we present BioCoder, a benchmark developed to evaluate existing pre-trained models in generating bioinformatics code. In relation to function-code generation, BioCoder covers potential package dependencies, class declarations, and global variables. It incorporates 1026 functions and 1243 methods in Python and Java from GitHub and 253 examples from the Rosalind Project. BioCoder incorporates a fuzz-testing framework for evaluation, and we have applied it to evaluate many models including InCoder, CodeGen, CodeGen2, SantaCoder, StarCoder, StarCoder+, InstructCodeT5+, and ChatGPT. Our detailed analysis of these models emphasizes the importance of domain knowledge, pragmatic code generation, and contextual understanding. Our dataset, benchmark, Docker images, and scripts required for testing are all available at https://github.com/gersteinlab/biocoder.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge