Jessica Gronsbell
PEHRT: A Common Pipeline for Harmonizing Electronic Health Record data for Translational Research
Sep 10, 2025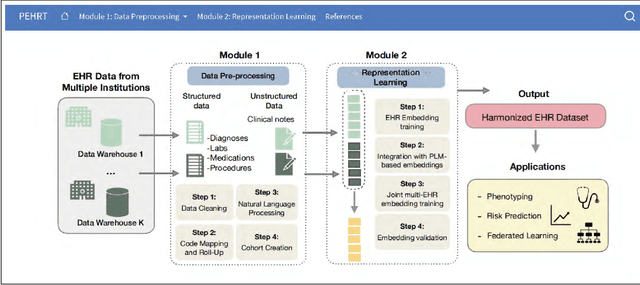
Abstract:Integrative analysis of multi-institutional Electronic Health Record (EHR) data enhances the reliability and generalizability of translational research by leveraging larger, more diverse patient cohorts and incorporating multiple data modalities. However, harmonizing EHR data across institutions poses major challenges due to data heterogeneity, semantic differences, and privacy concerns. To address these challenges, we introduce $\textit{PEHRT}$, a standardized pipeline for efficient EHR data harmonization consisting of two core modules: (1) data pre-processing and (2) representation learning. PEHRT maps EHR data to standard coding systems and uses advanced machine learning to generate research-ready datasets without requiring individual-level data sharing. Our pipeline is also data model agnostic and designed for streamlined execution across institutions based on our extensive real-world experience. We provide a complete suite of open source software, accompanied by a user-friendly tutorial, and demonstrate the utility of PEHRT in a variety of tasks using data from diverse healthcare systems.
fairmetrics: An R package for group fairness evaluation
Jun 06, 2025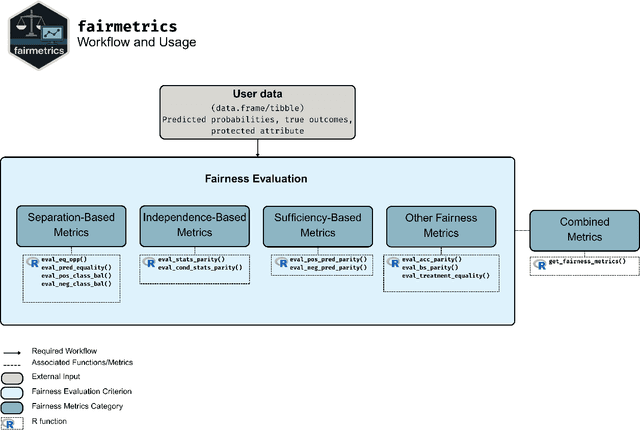
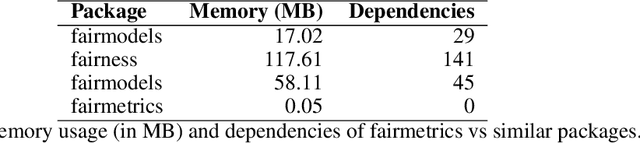
Abstract:Fairness is a growing area of machine learning (ML) that focuses on ensuring models do not produce systematically biased outcomes for specific groups, particularly those defined by protected attributes such as race, gender, or age. Evaluating fairness is a critical aspect of ML model development, as biased models can perpetuate structural inequalities. The {fairmetrics} R package offers a user-friendly framework for rigorously evaluating numerous group-based fairness criteria, including metrics based on independence (e.g., statistical parity), separation (e.g., equalized odds), and sufficiency (e.g., predictive parity). Group-based fairness criteria assess whether a model is equally accurate or well-calibrated across a set of predefined groups so that appropriate bias mitigation strategies can be implemented. {fairmetrics} provides both point and interval estimates for multiple metrics through a convenient wrapper function and includes an example dataset derived from the Medical Information Mart for Intensive Care, version II (MIMIC-II) database (Goldberger et al., 2000; Raffa, 2016).
Another look at inference after prediction
Nov 29, 2024


Abstract:Prediction-based (PB) inference is increasingly used in applications where the outcome of interest is difficult to obtain, but its predictors are readily available. Unlike traditional inference, PB inference performs statistical inference using a partially observed outcome and a set of covariates by leveraging a prediction of the outcome generated from a machine learning (ML) model. Motwani and Witten (2023) recently revisited two innovative PB inference approaches for ordinary least squares. They found that the method proposed by Wang et al. (2020) yields a consistent estimator for the association of interest when the ML model perfectly captures the underlying regression function. Conversely, the prediction-powered inference (PPI) method proposed by Angelopoulos et al. (2023) yields valid inference regardless of the model's accuracy. In this paper, we study the statistical efficiency of the PPI estimator. Our analysis reveals that a more efficient estimator, proposed 25 years ago by Chen and Chen (2000), can be obtained by simply adding a weight to the PPI estimator. We also contextualize PB inference with methods from the economics and statistics literature dating back to the 1960s. Our extensive theoretical and numerical analyses indicate that the Chen and Chen (CC) estimator offers a balance between robustness to ML model specification and statistical efficiency, making it the preferred choice for use in practice.
A tutorial on fairness in machine learning in healthcare
Jun 15, 2024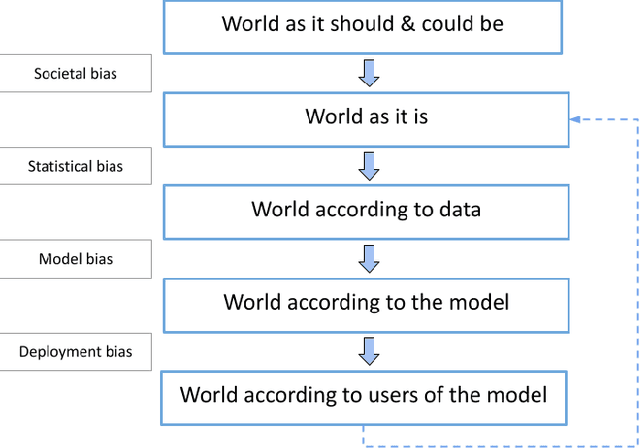
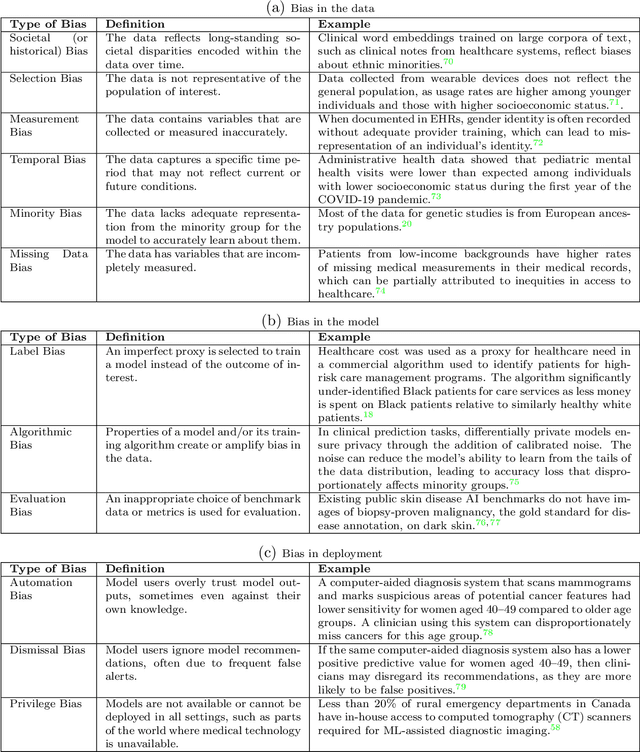
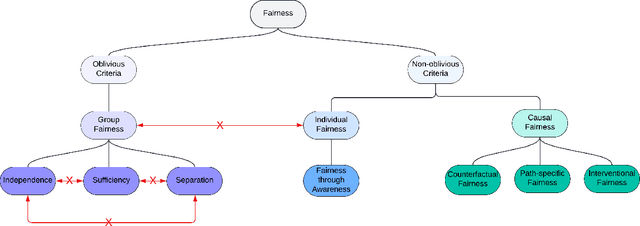
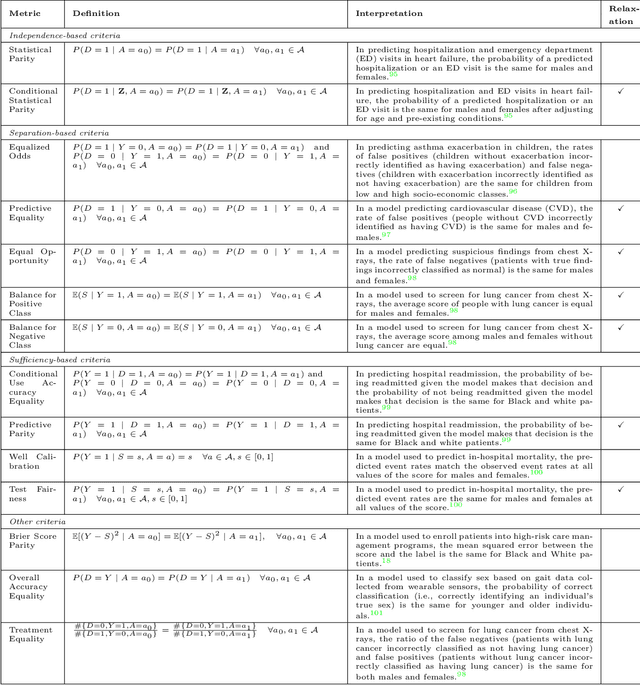
Abstract:$\textbf{OBJECTIVE}$: Ensuring that machine learning (ML) algorithms are safe and effective within all patient groups, and do not disadvantage particular patients, is essential to clinical decision making and preventing the reinforcement of existing healthcare inequities. The objective of this tutorial is to introduce the medical informatics community to the common notions of fairness within ML, focusing on clinical applications and implementation in practice. $\textbf{TARGET AUDIENCE}$: As gaps in fairness arise in a variety of healthcare applications, this tutorial is designed to provide an understanding of fairness, without assuming prior knowledge, to researchers and clinicians who make use of modern clinical data. $\textbf{SCOPE}$: We describe the fundamental concepts and methods used to define fairness in ML, including an overview of why models in healthcare may be unfair, a summary and comparison of the metrics used to quantify fairness, and a discussion of some ongoing research. We illustrate some of the fairness methods introduced through a case study of mortality prediction in a publicly available electronic health record dataset. Finally, we provide a user-friendly R package for comprehensive group fairness evaluation, enabling researchers and clinicians to assess fairness in their own ML work.
Efficient Estimation and Evaluation of Prediction Rules in Semi-Supervised Settings under Stratified Sampling
Oct 19, 2020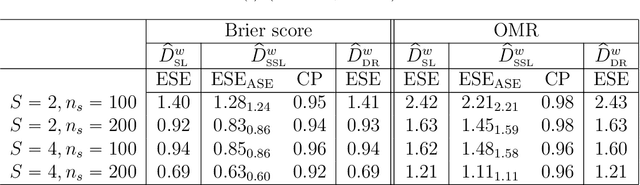
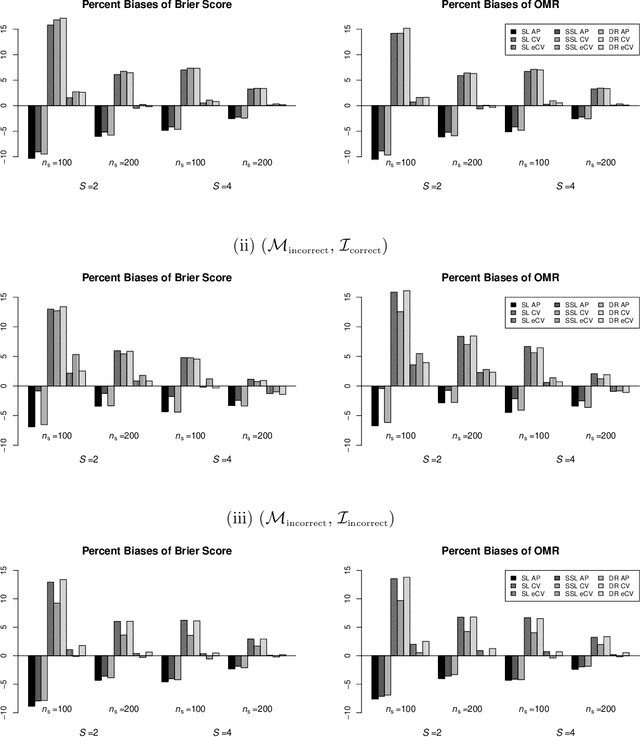
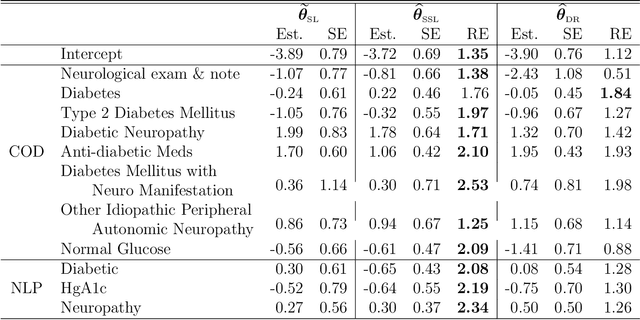
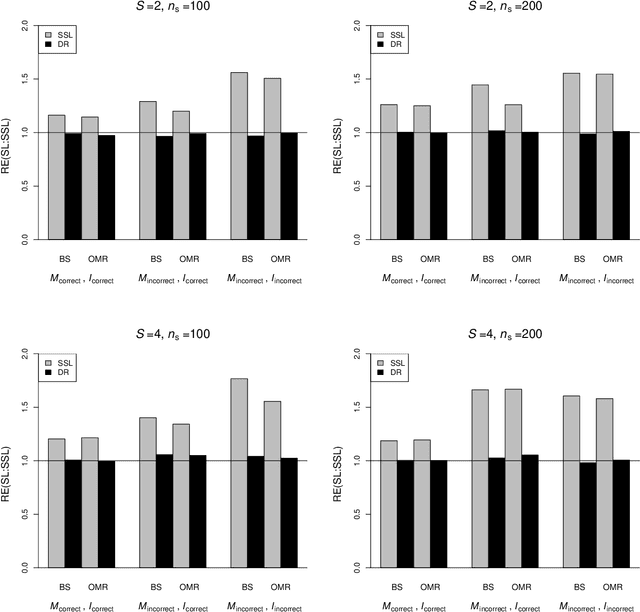
Abstract:In many contemporary applications, large amounts of unlabeled data are readily available while labeled examples are limited. There has been substantial interest in semi-supervised learning (SSL) which aims to leverage unlabeled data to improve estimation or prediction. However, current SSL literature focuses primarily on settings where labeled data is selected randomly from the population of interest. Non-random sampling, while posing additional analytical challenges, is highly applicable to many real world problems. Moreover, no SSL methods currently exist for estimating the prediction performance of a fitted model under non-random sampling. In this paper, we propose a two-step SSL procedure for evaluating a prediction rule derived from a working binary regression model based on the Brier score and overall misclassification rate under stratified sampling. In step I, we impute the missing labels via weighted regression with nonlinear basis functions to account for nonrandom sampling and to improve efficiency. In step II, we augment the initial imputations to ensure the consistency of the resulting estimators regardless of the specification of the prediction model or the imputation model. The final estimator is then obtained with the augmented imputations. We provide asymptotic theory and numerical studies illustrating that our proposals outperform their supervised counterparts in terms of efficiency gain. Our methods are motivated by electronic health records (EHR) research and validated with a real data analysis of an EHR-based study of diabetic neuropathy.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge