Jesper Holst Pedersen
Transfer learning for multi-center classification of chronic obstructive pulmonary disease
Nov 23, 2017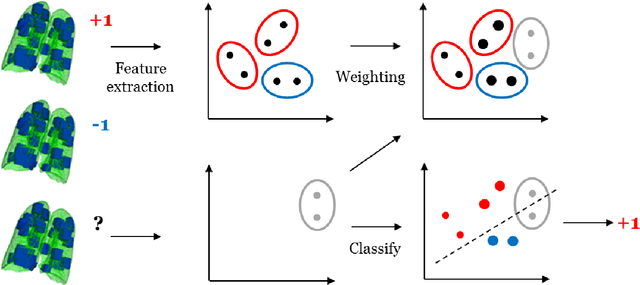
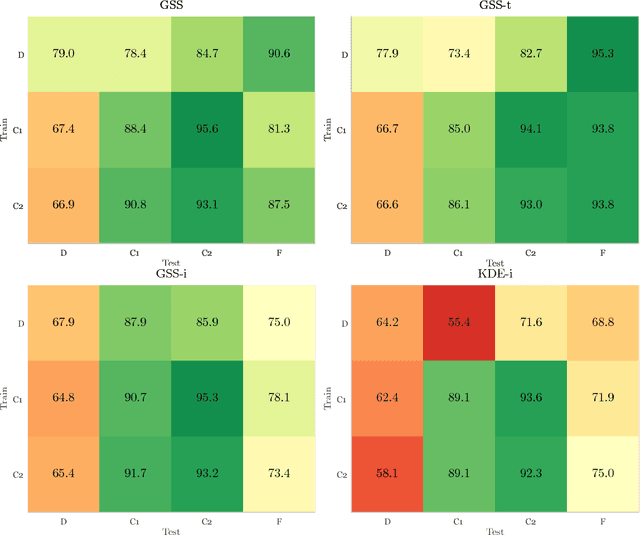
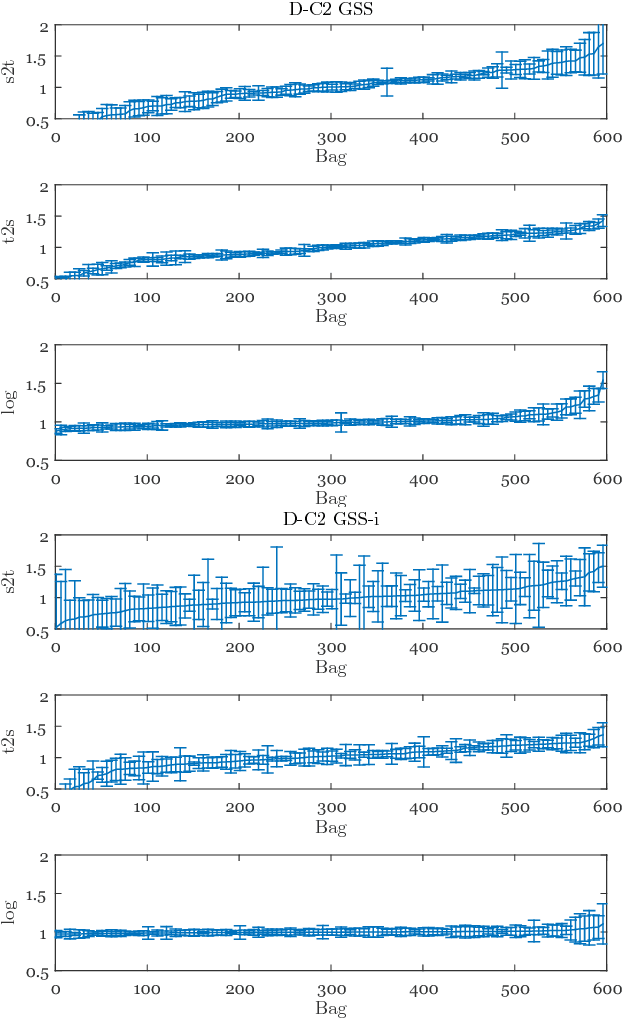
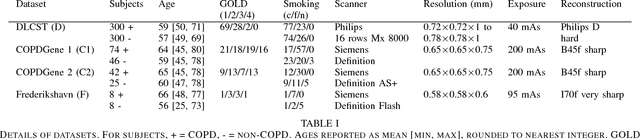
Abstract:Chronic obstructive pulmonary disease (COPD) is a lung disease which can be quantified using chest computed tomography (CT) scans. Recent studies have shown that COPD can be automatically diagnosed using weakly supervised learning of intensity and texture distributions. However, up till now such classifiers have only been evaluated on scans from a single domain, and it is unclear whether they would generalize across domains, such as different scanners or scanning protocols. To address this problem, we investigate classification of COPD in a multi-center dataset with a total of 803 scans from three different centers, four different scanners, with heterogenous subject distributions. Our method is based on Gaussian texture features, and a weighted logistic classifier, which increases the weights of samples similar to the test data. We show that Gaussian texture features outperform intensity features previously used in multi-center classification tasks. We also show that a weighting strategy based on a classifier that is trained to discriminate between scans from different domains, can further improve the results. To encourage further research into transfer learning methods for classification of COPD, upon acceptance of the paper we will release two feature datasets used in this study on http://bigr.nl/research/projects/copd
Classification of COPD with Multiple Instance Learning
Mar 15, 2017



Abstract:Chronic obstructive pulmonary disease (COPD) is a lung disease where early detection benefits the survival rate. COPD can be quantified by classifying patches of computed tomography images, and combining patch labels into an overall diagnosis for the image. As labeled patches are often not available, image labels are propagated to the patches, incorrectly labeling healthy patches in COPD patients as being affected by the disease. We approach quantification of COPD from lung images as a multiple instance learning (MIL) problem, which is more suitable for such weakly labeled data. We investigate various MIL assumptions in the context of COPD and show that although a concept region with COPD-related disease patterns is present, considering the whole distribution of lung tissue patches improves the performance. The best method is based on averaging instances and obtains an AUC of 0.742, which is higher than the previously reported best of 0.713 on the same dataset. Using the full training set further increases performance to 0.776, which is significantly higher (DeLong test) than previous results.
Geometric tree kernels: Classification of COPD from airway tree geometry
Apr 08, 2013



Abstract:Methodological contributions: This paper introduces a family of kernels for analyzing (anatomical) trees endowed with vector valued measurements made along the tree. While state-of-the-art graph and tree kernels use combinatorial tree/graph structure with discrete node and edge labels, the kernels presented in this paper can include geometric information such as branch shape, branch radius or other vector valued properties. In addition to being flexible in their ability to model different types of attributes, the presented kernels are computationally efficient and some of them can easily be computed for large datasets (N of the order 10.000) of trees with 30-600 branches. Combining the kernels with standard machine learning tools enables us to analyze the relation between disease and anatomical tree structure and geometry. Experimental results: The kernels are used to compare airway trees segmented from low-dose CT, endowed with branch shape descriptors and airway wall area percentage measurements made along the tree. Using kernelized hypothesis testing we show that the geometric airway trees are significantly differently distributed in patients with Chronic Obstructive Pulmonary Disease (COPD) than in healthy individuals. The geometric tree kernels also give a significant increase in the classification accuracy of COPD from geometric tree structure endowed with airway wall thickness measurements in comparison with state-of-the-art methods, giving further insight into the relationship between airway wall thickness and COPD. Software: Software for computing kernels and statistical tests is available at http://image.diku.dk/aasa/software.php.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge