Jeremy Johnson
High Rank Matrix Completion via Grassmannian Proxy Fusion
Jan 30, 2026Abstract:This paper approaches high-rank matrix completion (HRMC) by filling missing entries in a data matrix where columns lie near a union of subspaces, clustering these columns, and identifying the underlying subspaces. Current methods often lack theoretical support, produce uninterpretable results, and require more samples than theoretically necessary. We propose clustering incomplete vectors by grouping proxy subspaces and minimizing two criteria over the Grassmannian: (a) the chordal distance between each point and its corresponding subspace and (b) the geodesic distances between subspaces of all data points. Experiments on synthetic and real datasets demonstrate that our method performs comparably to leading methods in high sampling rates and significantly better in low sampling rates, thus narrowing the gap to the theoretical sampling limit of HRMC.
End-to-end learning of brain tissue segmentation from imperfect labeling
Jun 05, 2017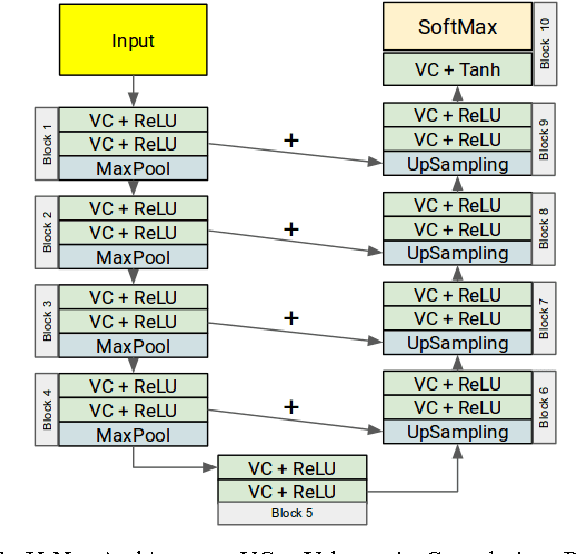
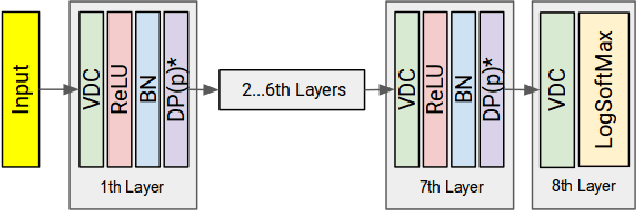
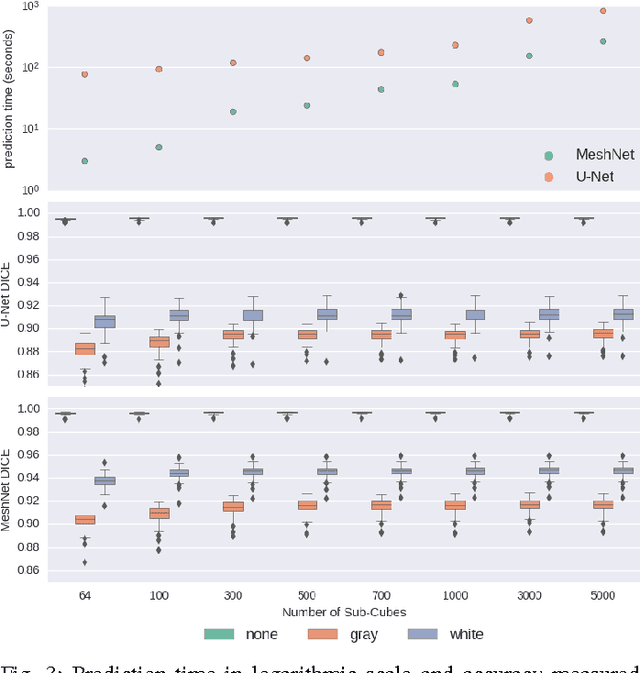
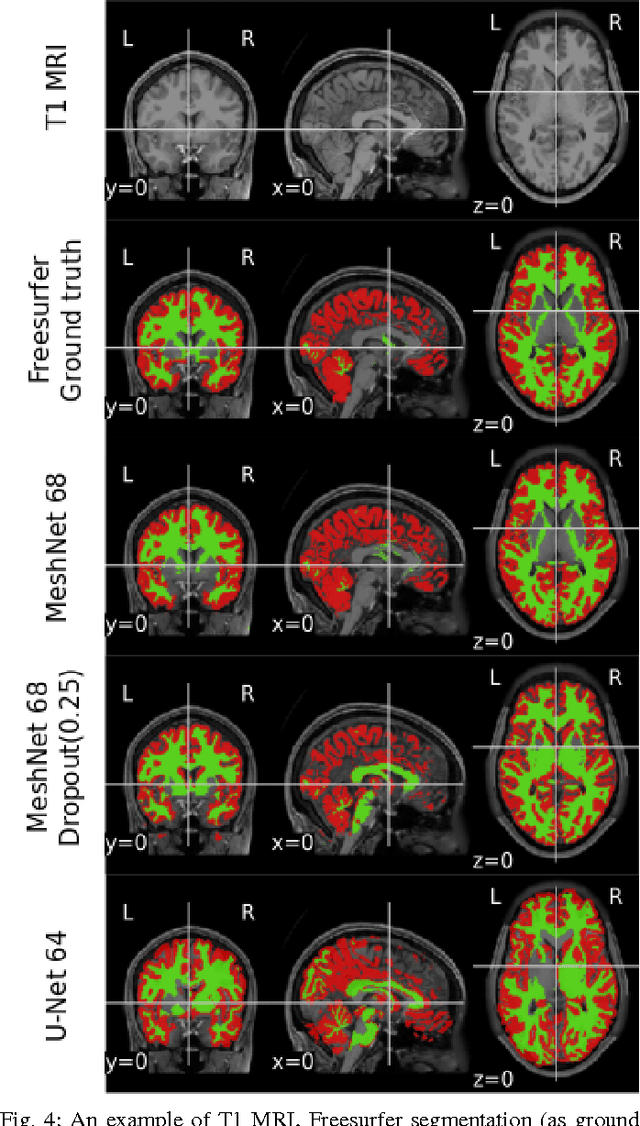
Abstract:Segmenting a structural magnetic resonance imaging (MRI) scan is an important pre-processing step for analytic procedures and subsequent inferences about longitudinal tissue changes. Manual segmentation defines the current gold standard in quality but is prohibitively expensive. Automatic approaches are computationally intensive, incredibly slow at scale, and error prone due to usually involving many potentially faulty intermediate steps. In order to streamline the segmentation, we introduce a deep learning model that is based on volumetric dilated convolutions, subsequently reducing both processing time and errors. Compared to its competitors, the model has a reduced set of parameters and thus is easier to train and much faster to execute. The contrast in performance between the dilated network and its competitors becomes obvious when both are tested on a large dataset of unprocessed human brain volumes. The dilated network consistently outperforms not only another state-of-the-art deep learning approach, the up convolutional network, but also the ground truth on which it was trained. Not only can the incredible speed of our model make large scale analyses much easier but we also believe it has great potential in a clinical setting where, with little to no substantial delay, a patient and provider can go over test results.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge