Jeff Guo
KL-Regularized Reinforcement Learning is Designed to Mode Collapse
Oct 23, 2025Abstract:It is commonly believed that optimizing the reverse KL divergence results in "mode seeking", while optimizing forward KL results in "mass covering", with the latter being preferred if the goal is to sample from multiple diverse modes. We show -- mathematically and empirically -- that this intuition does not necessarily transfer well to doing reinforcement learning with reverse/forward KL regularization (e.g. as commonly used with language models). Instead, the choice of reverse/forward KL determines the family of optimal target distributions, parameterized by the regularization coefficient. Mode coverage depends primarily on other factors, such as regularization strength, and relative scales between rewards and reference probabilities. Further, we show commonly used settings such as low regularization strength and equal verifiable rewards tend to specify unimodal target distributions, meaning the optimization objective is, by construction, non-diverse. We leverage these insights to construct a simple, scalable, and theoretically justified algorithm. It makes minimal changes to reward magnitudes, yet optimizes for a target distribution which puts high probability over all high-quality sampling modes. In experiments, this simple modification works to post-train both Large Language Models and Chemical Language Models to have higher solution quality and diversity, without any external signals of diversity, and works with both forward and reverse KL when using either naively fails.
Generative Molecular Design with Steerable and Granular Synthesizability Control
May 13, 2025Abstract:Synthesizability in small molecule generative design remains a bottleneck. Existing works that do consider synthesizability can output predicted synthesis routes for generated molecules. However, there has been minimal attention in addressing the ease of synthesis and enabling flexibility to incorporate desired reaction constraints. In this work, we propose a small molecule generative design framework that enables steerable and granular synthesizability control. Generated molecules satisfy arbitrary multi-parameter optimization objectives with predicted synthesis routes containing pre-defined allowed reactions, while optionally avoiding others. One can also enforce that all reactions belong to a pre-defined set. We show the capability to mix-and-match these reaction constraints across the most common medicinal chemistry transformations. Next, we show how our framework can be used to valorize industrial byproducts towards de novo optimized molecules. Going further, we demonstrate how granular control over synthesizability constraints can loosely mimic virtual screening of ultra-large make-on-demand libraries. Using only a single GPU, we generate and dock 15k molecules to identify promising candidates in Freedom 4.0 constituting 142B make-on-demand molecules (assessing only 0.00001% of the library). Generated molecules satisfying the reaction constraints have > 90% exact match rate. Lastly, we benchmark our framework against recent synthesizability-constrained generative models and demonstrate the highest sample efficiency even when imposing the additional constraint that all molecules must be synthesizable from a single reaction type. The main theme is demonstrating that a pre-trained generalist molecular generative model can be incentivized to generate property-optimized small molecules under challenging synthesizability constraints through reinforcement learning.
LLM-Augmented Chemical Synthesis and Design Decision Programs
May 11, 2025Abstract:Retrosynthesis, the process of breaking down a target molecule into simpler precursors through a series of valid reactions, stands at the core of organic chemistry and drug development. Although recent machine learning (ML) research has advanced single-step retrosynthetic modeling and subsequent route searches, these solutions remain restricted by the extensive combinatorial space of possible pathways. Concurrently, large language models (LLMs) have exhibited remarkable chemical knowledge, hinting at their potential to tackle complex decision-making tasks in chemistry. In this work, we explore whether LLMs can successfully navigate the highly constrained, multi-step retrosynthesis planning problem. We introduce an efficient scheme for encoding reaction pathways and present a new route-level search strategy, moving beyond the conventional step-by-step reactant prediction. Through comprehensive evaluations, we show that our LLM-augmented approach excels at retrosynthesis planning and extends naturally to the broader challenge of synthesizable molecular design.
Tango*: Constrained synthesis planning using chemically informed value functions
Dec 04, 2024Abstract:Computer-aided synthesis planning (CASP) has made significant strides in generating retrosynthetic pathways for simple molecules in a non-constrained fashion. Recent work introduces a specialised bidirectional search algorithm with forward and retro expansion to address the starting material-constrained synthesis problem, allowing CASP systems to provide synthesis pathways from specified starting materials, such as waste products or renewable feed-stocks. In this work, we introduce a simple guided search which allows solving the starting material-constrained synthesis planning problem using an existing, uni-directional search algorithm, Retro*. We show that by optimising a single hyperparameter, Tango* outperforms existing methods in terms of efficiency and solve rate. We find the Tango* cost function catalyses strong improvements for the bidirectional DESP methods. Our method also achieves lower wall clock times while proposing synthetic routes of similar length, a common metric for route quality. Finally, we highlight potential reasons for the strong performance of Tango over neural guided search methods
It Takes Two to Tango: Directly Optimizing for Constrained Synthesizability in Generative Molecular Design
Oct 15, 2024Abstract:Constrained synthesizability is an unaddressed challenge in generative molecular design. In particular, designing molecules satisfying multi-parameter optimization objectives, while simultaneously being synthesizable and enforcing the presence of specific commercial building blocks in the synthesis. This is practically important for molecule re-purposing, sustainability, and efficiency. In this work, we propose a novel reward function called TANimoto Group Overlap (TANGO), which uses chemistry principles to transform a sparse reward function into a dense and learnable reward function -- crucial for reinforcement learning. TANGO can augment general-purpose molecular generative models to directly optimize for constrained synthesizability while simultaneously optimizing for other properties relevant to drug discovery using reinforcement learning. Our framework is general and addresses starting-material, intermediate, and divergent synthesis constraints. Contrary to most existing works in the field, we show that incentivizing a general-purpose (without any inductive biases) model is a productive approach to navigating challenging optimization scenarios. We demonstrate this by showing that the trained models explicitly learn a desirable distribution. Our framework is the first generative approach to tackle constrained synthesizability.
Saturn: Sample-efficient Generative Molecular Design using Memory Manipulation
May 27, 2024Abstract:Generative molecular design for drug discovery has very recently achieved a wave of experimental validation, with language-based backbones being the most common architectures employed. The most important factor for downstream success is whether an in silico oracle is well correlated with the desired end-point. To this end, current methods use cheaper proxy oracles with higher throughput before evaluating the most promising subset with high-fidelity oracles. The ability to directly optimize high-fidelity oracles would greatly enhance generative design and be expected to improve hit rates. However, current models are not efficient enough to consider such a prospect, exemplifying the sample efficiency problem. In this work, we introduce Saturn, which leverages the Augmented Memory algorithm and demonstrates the first application of the Mamba architecture for generative molecular design. We elucidate how experience replay with data augmentation improves sample efficiency and how Mamba synergistically exploits this mechanism. Saturn outperforms 22 models on multi-parameter optimization tasks relevant to drug discovery and may possess sufficient sample efficiency to consider the prospect of directly optimizing high-fidelity oracles.
Beam Enumeration: Probabilistic Explainability For Sample Efficient Self-conditioned Molecular Design
Sep 25, 2023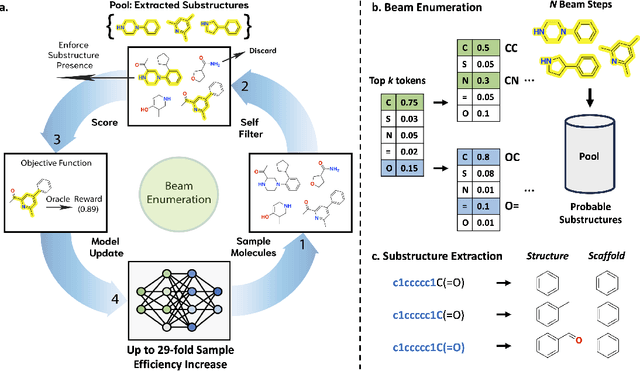
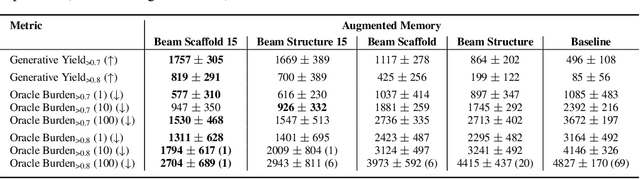
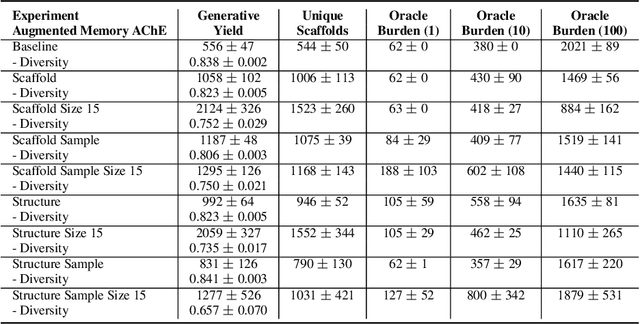
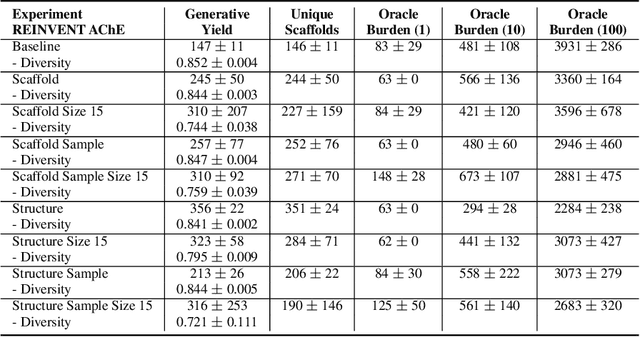
Abstract:Generative molecular design has moved from proof-of-concept to real-world applicability, as marked by the surge in very recent papers reporting experimental validation. Key challenges in explainability and sample efficiency present opportunities to enhance generative design to directly optimize expensive high-fidelity oracles and provide actionable insights to domain experts. Here, we propose Beam Enumeration to exhaustively enumerate the most probable sub-sequences from language-based molecular generative models and show that molecular substructures can be extracted. When coupled with reinforcement learning, extracted substructures become meaningful, providing a source of explainability and improving sample efficiency through self-conditioned generation. Beam Enumeration is generally applicable to any language-based molecular generative model and notably further improves the performance of the recently reported Augmented Memory algorithm, which achieved the new state-of-the-art on the Practical Molecular Optimization benchmark for sample efficiency. The combined algorithm generates more high reward molecules and faster, given a fixed oracle budget. Beam Enumeration is the first method to jointly address explainability and sample efficiency for molecular design.
Augmented Memory: Capitalizing on Experience Replay to Accelerate De Novo Molecular Design
May 10, 2023Abstract:Sample efficiency is a fundamental challenge in de novo molecular design. Ideally, molecular generative models should learn to satisfy a desired objective under minimal oracle evaluations (computational prediction or wet-lab experiment). This problem becomes more apparent when using oracles that can provide increased predictive accuracy but impose a significant cost. Consequently, these oracles cannot be directly optimized under a practical budget. Molecular generative models have shown remarkable sample efficiency when coupled with reinforcement learning, as demonstrated in the Practical Molecular Optimization (PMO) benchmark. Here, we propose a novel algorithm called Augmented Memory that combines data augmentation with experience replay. We show that scores obtained from oracle calls can be reused to update the model multiple times. We compare Augmented Memory to previously proposed algorithms and show significantly enhanced sample efficiency in an exploitation task and a drug discovery case study requiring both exploration and exploitation. Our method achieves a new state-of-the-art in the PMO benchmark which enforces a computational budget, outperforming the previous best performing method on 19/23 tasks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge