Ivor J. A. Simpson
Learning Mechanistic Subtypes of Neurodegeneration with a Physics-Informed Variational Autoencoder Mixture Model
Sep 18, 2025Abstract:Modelling the underlying mechanisms of neurodegenerative diseases demands methods that capture heterogeneous and spatially varying dynamics from sparse, high-dimensional neuroimaging data. Integrating partial differential equation (PDE) based physics knowledge with machine learning provides enhanced interpretability and utility over classic numerical methods. However, current physics-integrated machine learning methods are limited to considering a single PDE, severely limiting their application to diseases where multiple mechanisms are responsible for different groups (i.e., subtypes) and aggravating problems with model misspecification and degeneracy. Here, we present a deep generative model for learning mixtures of latent dynamic models governed by physics-based PDEs, going beyond traditional approaches that assume a single PDE structure. Our method integrates reaction-diffusion PDEs within a variational autoencoder (VAE) mixture model framework, supporting inference of subtypes of interpretable latent variables (e.g. diffusivity and reaction rates) from neuroimaging data. We evaluate our method on synthetic benchmarks and demonstrate its potential for uncovering mechanistic subtypes of Alzheimer's disease progression from positron emission tomography (PET) data.
Investigating the Role of Bilateral Symmetry for Inpainting Brain MRI
Apr 14, 2025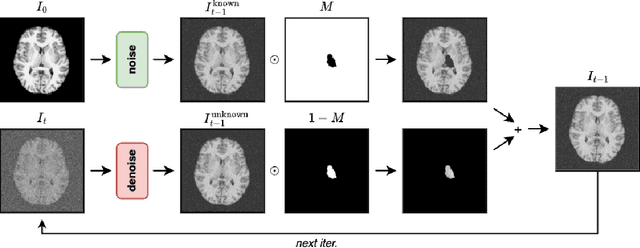
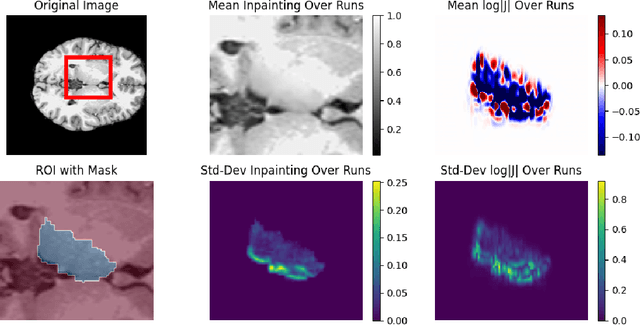
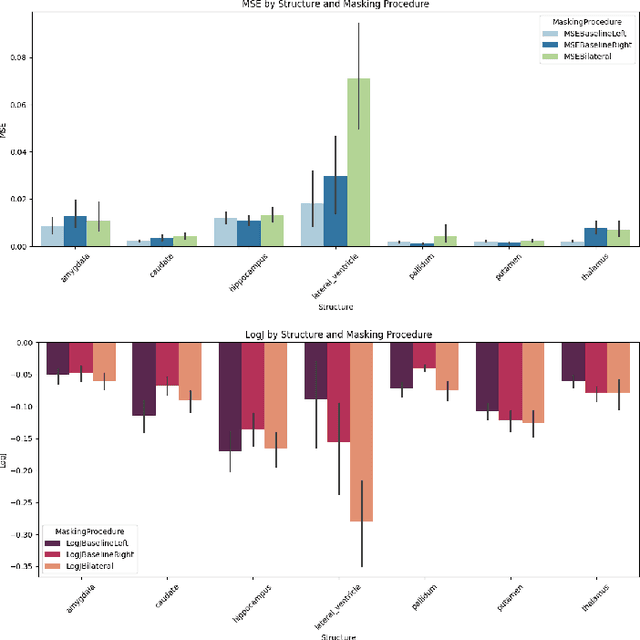
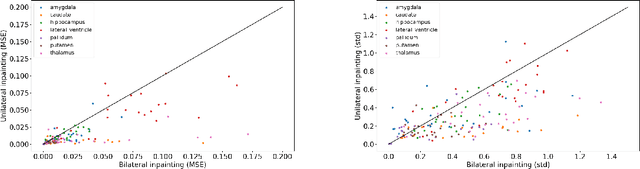
Abstract:Inpainting has recently emerged as a valuable and interesting technology to employ in the analysis of medical imaging data, in particular brain MRI. A wide variety of methodologies for inpainting MRI have been proposed and demonstrated on tasks including anomaly detection. In this work we investigate the statistical relationship between inpainted brain structures and the amount of subject-specific conditioning information, i.e. the other areas of the image that are masked. In particular, we analyse the distribution of inpainting results when masking additional regions of the image, specifically the contra-lateral structure. This allows us to elucidate where in the brain the model is drawing information from, and in particular, what is the importance of hemispherical symmetry? Our experiments interrogate a diffusion inpainting model through analysing the inpainting of subcortical brain structures based on intensity and estimated area change. We demonstrate that some structures show a strong influence of symmetry in the conditioning of the inpainting process.
Capturing Longitudinal Changes in Brain Morphology Using Temporally Parameterized Neural Displacement Fields
Apr 13, 2025Abstract:Longitudinal image registration enables studying temporal changes in brain morphology which is useful in applications where monitoring the growth or atrophy of specific structures is important. However this task is challenging due to; noise/artifacts in the data and quantifying small anatomical changes between sequential scans. We propose a novel longitudinal registration method that models structural changes using temporally parameterized neural displacement fields. Specifically, we implement an implicit neural representation (INR) using a multi-layer perceptron that serves as a continuous coordinate-based approximation of the deformation field at any time point. In effect, for any N scans of a particular subject, our model takes as input a 3D spatial coordinate location x, y, z and a corresponding temporal representation t and learns to describe the continuous morphology of structures for both observed and unobserved points in time. Furthermore, we leverage the analytic derivatives of the INR to derive a new regularization function that enforces monotonic rate of change in the trajectory of the voxels, which is shown to provide more biologically plausible patterns. We demonstrate the effectiveness of our method on 4D brain MR registration.
HyperPredict: Estimating Hyperparameter Effects for Instance-Specific Regularization in Deformable Image Registration
Mar 04, 2024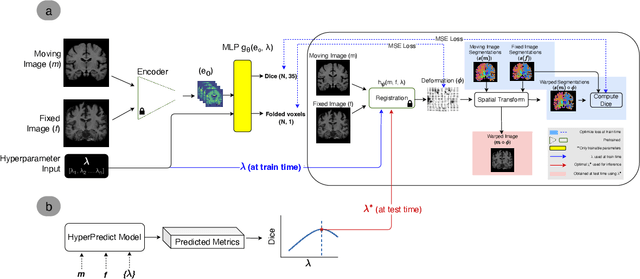

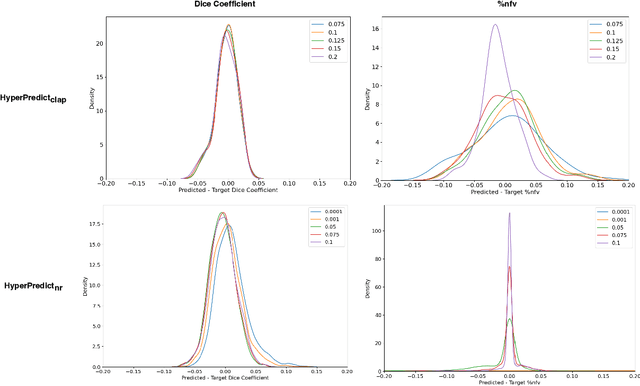
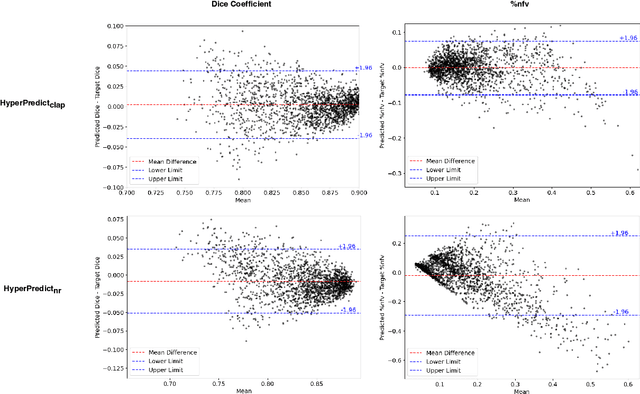
Abstract:Methods for medical image registration infer geometric transformations that align pairs/groups of images by maximising an image similarity metric. This problem is ill-posed as several solutions may have equivalent likelihoods, also optimising purely for image similarity can yield implausible transformations. For these reasons regularization terms are essential to obtain meaningful registration results. However, this requires the introduction of at least one hyperparameter often termed {\lambda}, that serves as a tradeoff between loss terms. In some situations, the quality of the estimated transformation greatly depends on hyperparameter choice, and different choices may be required depending on the characteristics of the data. Analyzing the effect of these hyperparameters requires labelled data, which is not commonly available at test-time. In this paper, we propose a method for evaluating the influence of hyperparameters and subsequently selecting an optimal value for given image pairs. Our approach which we call HyperPredict, implements a Multi-Layer Perceptron that learns the effect of selecting particular hyperparameters for registering an image pair by predicting the resulting segmentation overlap and measure of deformation smoothness. This approach enables us to select optimal hyperparameters at test time without requiring labelled data, removing the need for a one-size-fits-all cross-validation approach. Furthermore, the criteria used to define optimal hyperparameter is flexible post-training, allowing us to efficiently choose specific properties. We evaluate our proposed method on the OASIS brain MR dataset using a recent deep learning approach(cLapIRN) and an algorithmic method(Niftyreg). Our results demonstrate good performance in predicting the effects of regularization hyperparameters and highlight the benefits of our image-pair specific approach to hyperparameter selection.
Compressed Sensing MRI Reconstruction Regularized by VAEs with Structured Image Covariance
Oct 26, 2022Abstract:Learned regularization for MRI reconstruction can provide complex data-driven priors to inverse problems while still retaining the control and insight of a variational regularization method. Moreover, unsupervised learning, without paired training data, allows the learned regularizer to remain flexible to changes in the forward problem such as noise level, sampling pattern or coil sensitivities. One such approach uses generative models, trained on ground-truth images, as priors for inverse problems, penalizing reconstructions far from images the generator can produce. In this work, we utilize variational autoencoders (VAEs) that generate not only an image but also a covariance uncertainty matrix for each image. The covariance can model changing uncertainty dependencies caused by structure in the image, such as edges or objects, and provides a new distance metric from the manifold of learned images. We demonstrate these novel generative regularizers on radially sub-sampled MRI knee measurements from the fastMRI dataset and compare them to other unlearned, unsupervised and supervised methods. Our results show that the proposed method is competitive with other state-of-the-art methods and behaves consistently with changing sampling patterns and noise levels.
Flexible Amortized Variational Inference in qBOLD MRI
Apr 07, 2022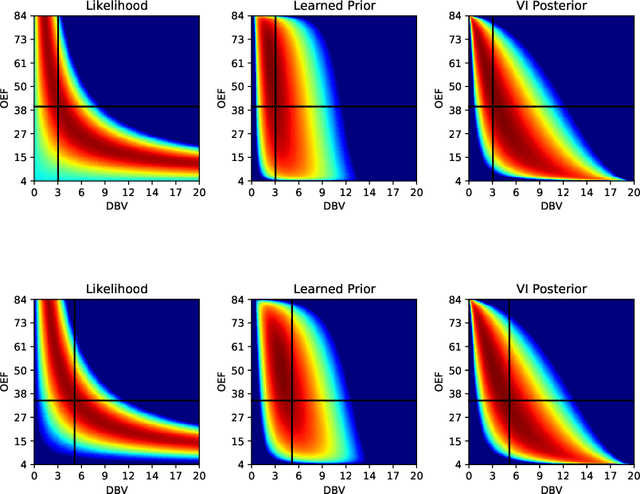
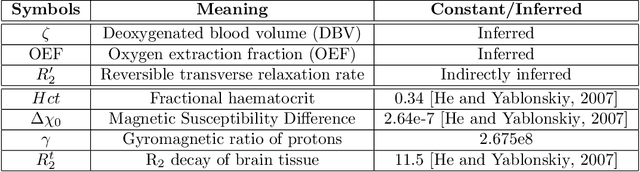
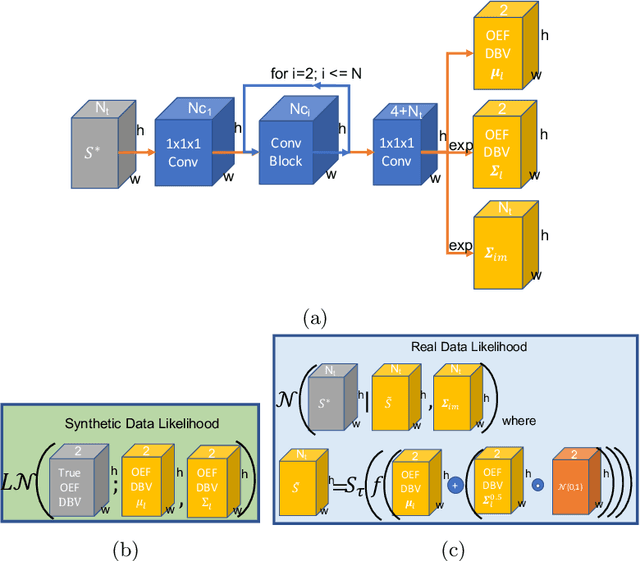
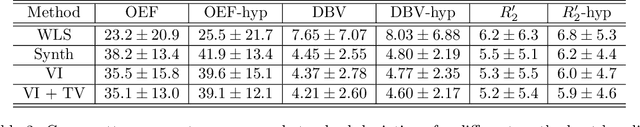
Abstract:Streamlined qBOLD acquisitions enable experimentally straightforward observations of brain oxygen metabolism. $R_2^\prime$ maps are easily inferred; however, the Oxygen extraction fraction (OEF) and deoxygenated blood volume (DBV) are more ambiguously determined from the data. As such, existing inference methods tend to yield very noisy and underestimated OEF maps, while overestimating DBV. This work describes a novel probabilistic machine learning approach that can infer plausible distributions of OEF and DBV. Initially, we create a model that produces informative voxelwise prior distribution based on synthetic training data. Contrary to prior work, we model the joint distribution of OEF and DBV through a scaled multivariate logit-Normal distribution, which enables the values to be constrained within a plausible range. The prior distribution model is used to train an efficient amortized variational Bayesian inference model. This model learns to infer OEF and DBV by predicting real image data, with few training data required, using the signal equations as a forward model. We demonstrate that our approach enables the inference of smooth OEF and DBV maps, with a physiologically plausible distribution that can be adapted through specification of an informative prior distribution. Other benefits include model comparison (via the evidence lower bound) and uncertainty quantification for identifying image artefacts. Results are demonstrated on a small study comparing subjects undergoing hyperventilation and at rest. We illustrate that the proposed approach allows measurement of gray matter differences in OEF and DBV and enables voxelwise comparison between conditions, where we observe significant increases in OEF and $R_2^\prime$ during hyperventilation.
Learning Structured Gaussians to Approximate Deep Ensembles
Mar 29, 2022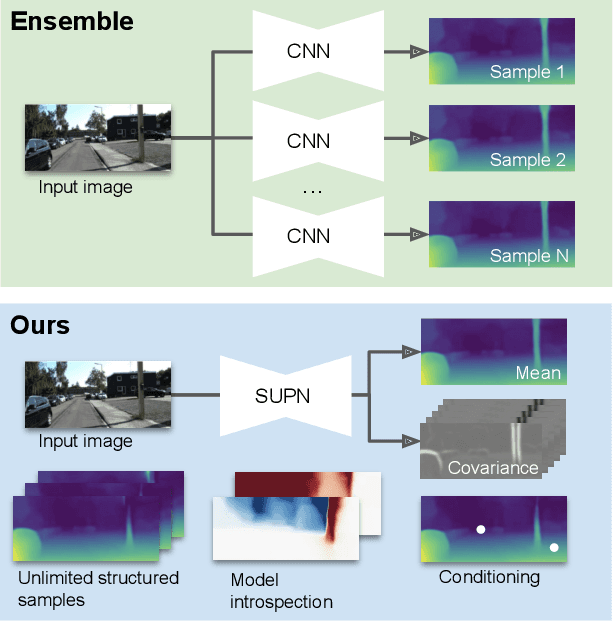
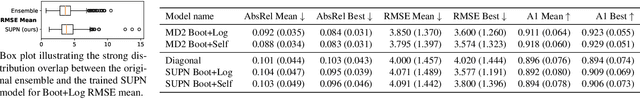
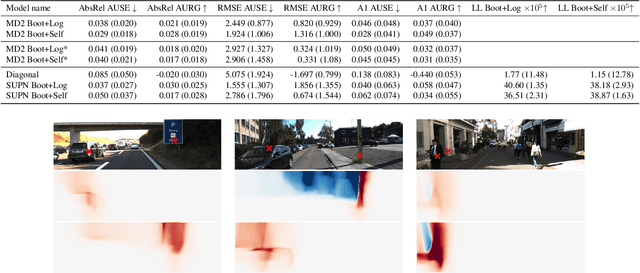
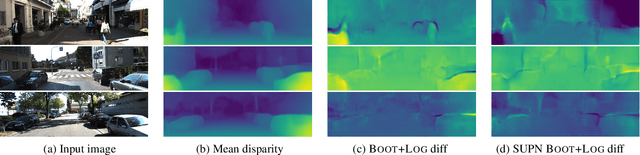
Abstract:This paper proposes using a sparse-structured multivariate Gaussian to provide a closed-form approximator for the output of probabilistic ensemble models used for dense image prediction tasks. This is achieved through a convolutional neural network that predicts the mean and covariance of the distribution, where the inverse covariance is parameterised by a sparsely structured Cholesky matrix. Similarly to distillation approaches, our single network is trained to maximise the probability of samples from pre-trained probabilistic models, in this work we use a fixed ensemble of networks. Once trained, our compact representation can be used to efficiently draw spatially correlated samples from the approximated output distribution. Importantly, this approach captures the uncertainty and structured correlations in the predictions explicitly in a formal distribution, rather than implicitly through sampling alone. This allows direct introspection of the model, enabling visualisation of the learned structure. Moreover, this formulation provides two further benefits: estimation of a sample probability, and the introduction of arbitrary spatial conditioning at test time. We demonstrate the merits of our approach on monocular depth estimation and show that the advantages of our approach are obtained with comparable quantitative performance.
Estimation of Large Motion in Lung CT by Integrating Regularized Keypoint Correspondences into Dense Deformable Registration
Jul 02, 2018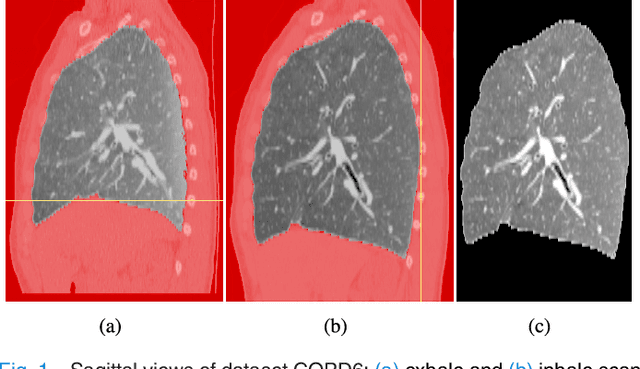
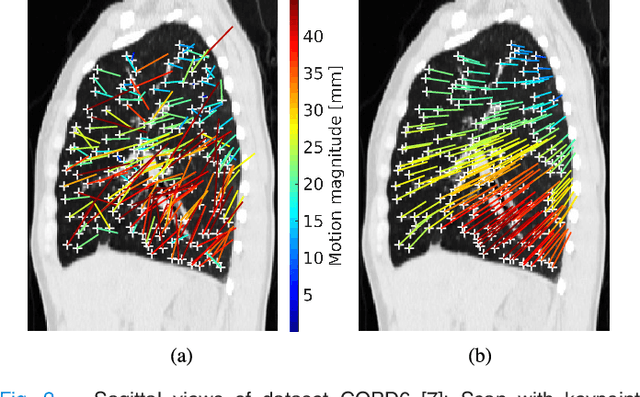
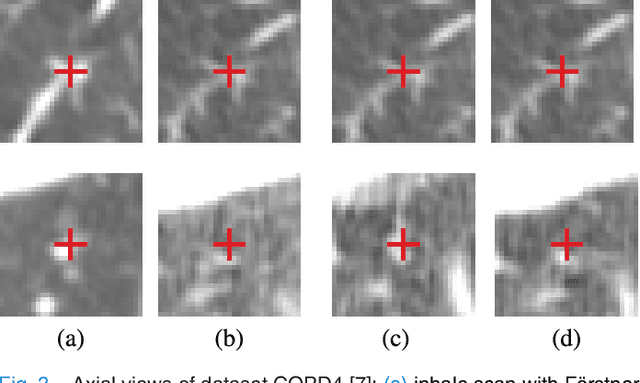
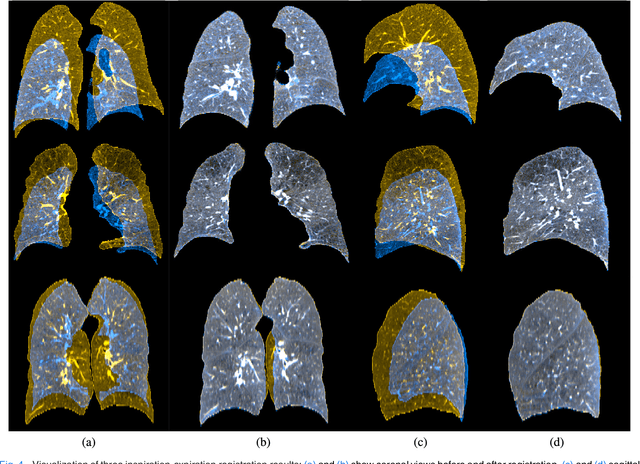
Abstract:We present a novel algorithm for the registration of pulmonary CT scans. Our method is designed for large respiratory motion by integrating sparse keypoint correspondences into a dense continuous optimization framework. The detection of keypoint correspondences enables robustness against large deformations by jointly optimizing over a large number of potential discrete displacements, whereas the dense continuous registration achieves subvoxel alignment with smooth transformations. Both steps are driven by the same normalized gradient fields data term. We employ curvature regularization and a volume change control mechanism to prevent foldings of the deformation grid and restrict the determinant of the Jacobian to physiologically meaningful values. Keypoint correspondences are integrated into the dense registration by a quadratic penalty with adaptively determined weight. Using a parallel matrix-free derivative calculation scheme, a runtime of about 5 min was realized on a standard PC. The proposed algorithm ranks first in the EMPIRE10 challenge on pulmonary image registration. Moreover, it achieves an average landmark distance of 0.82 mm on the DIR-Lab COPD database, thereby improving upon the state of the art in accuracy by 15%. Our algorithm is the first to reach the inter-observer variability in landmark annotation on this dataset.
* 12 pages, 7 figures, \c{opyright} 2017 IEEE. Personal use is permitted, but republication/redistribution requires IEEE permission
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge