Flexible Amortized Variational Inference in qBOLD MRI
Paper and Code
Apr 07, 2022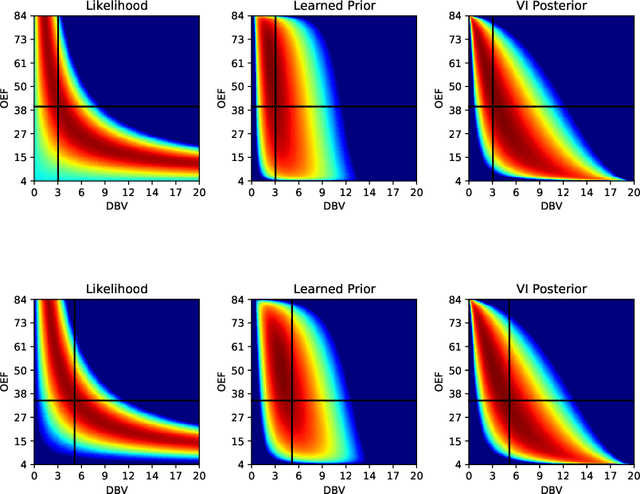
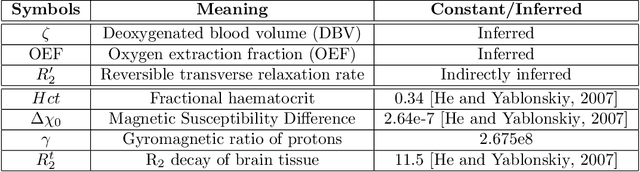
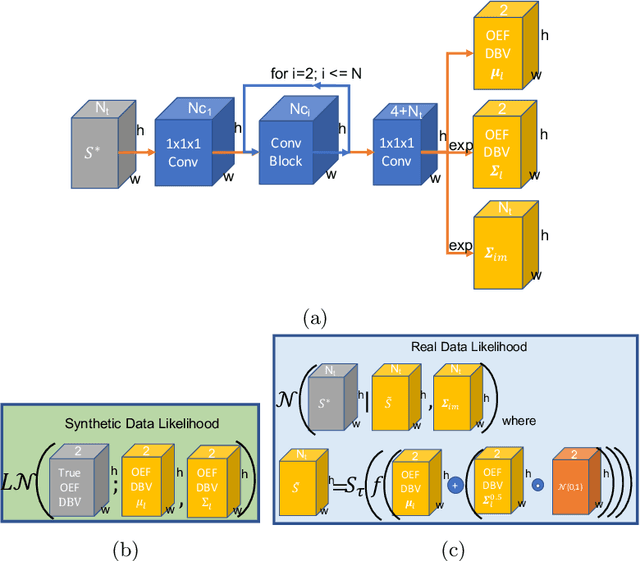
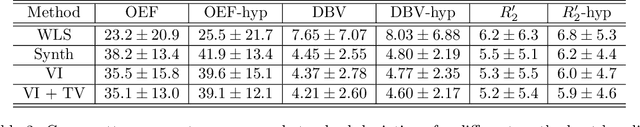
Streamlined qBOLD acquisitions enable experimentally straightforward observations of brain oxygen metabolism. $R_2^\prime$ maps are easily inferred; however, the Oxygen extraction fraction (OEF) and deoxygenated blood volume (DBV) are more ambiguously determined from the data. As such, existing inference methods tend to yield very noisy and underestimated OEF maps, while overestimating DBV. This work describes a novel probabilistic machine learning approach that can infer plausible distributions of OEF and DBV. Initially, we create a model that produces informative voxelwise prior distribution based on synthetic training data. Contrary to prior work, we model the joint distribution of OEF and DBV through a scaled multivariate logit-Normal distribution, which enables the values to be constrained within a plausible range. The prior distribution model is used to train an efficient amortized variational Bayesian inference model. This model learns to infer OEF and DBV by predicting real image data, with few training data required, using the signal equations as a forward model. We demonstrate that our approach enables the inference of smooth OEF and DBV maps, with a physiologically plausible distribution that can be adapted through specification of an informative prior distribution. Other benefits include model comparison (via the evidence lower bound) and uncertainty quantification for identifying image artefacts. Results are demonstrated on a small study comparing subjects undergoing hyperventilation and at rest. We illustrate that the proposed approach allows measurement of gray matter differences in OEF and DBV and enables voxelwise comparison between conditions, where we observe significant increases in OEF and $R_2^\prime$ during hyperventilation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge