Peter A. Wijeratne
Learning Mechanistic Subtypes of Neurodegeneration with a Physics-Informed Variational Autoencoder Mixture Model
Sep 18, 2025Abstract:Modelling the underlying mechanisms of neurodegenerative diseases demands methods that capture heterogeneous and spatially varying dynamics from sparse, high-dimensional neuroimaging data. Integrating partial differential equation (PDE) based physics knowledge with machine learning provides enhanced interpretability and utility over classic numerical methods. However, current physics-integrated machine learning methods are limited to considering a single PDE, severely limiting their application to diseases where multiple mechanisms are responsible for different groups (i.e., subtypes) and aggravating problems with model misspecification and degeneracy. Here, we present a deep generative model for learning mixtures of latent dynamic models governed by physics-based PDEs, going beyond traditional approaches that assume a single PDE structure. Our method integrates reaction-diffusion PDEs within a variational autoencoder (VAE) mixture model framework, supporting inference of subtypes of interpretable latent variables (e.g. diffusivity and reaction rates) from neuroimaging data. We evaluate our method on synthetic benchmarks and demonstrate its potential for uncovering mechanistic subtypes of Alzheimer's disease progression from positron emission tomography (PET) data.
Investigating the Role of Bilateral Symmetry for Inpainting Brain MRI
Apr 14, 2025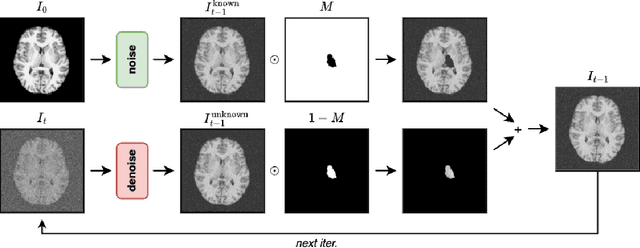
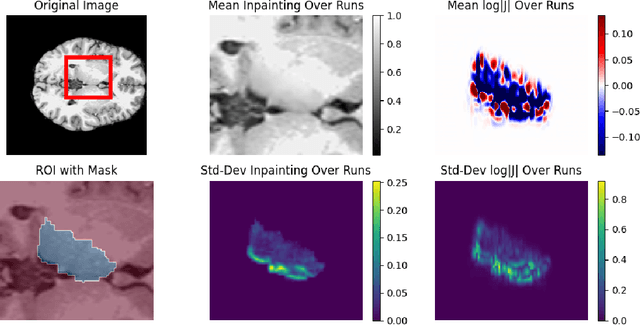
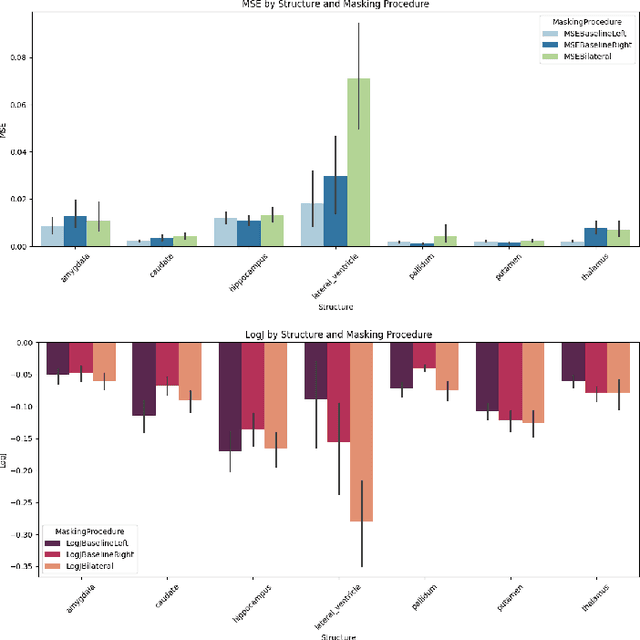
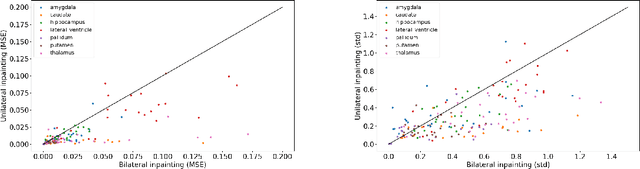
Abstract:Inpainting has recently emerged as a valuable and interesting technology to employ in the analysis of medical imaging data, in particular brain MRI. A wide variety of methodologies for inpainting MRI have been proposed and demonstrated on tasks including anomaly detection. In this work we investigate the statistical relationship between inpainted brain structures and the amount of subject-specific conditioning information, i.e. the other areas of the image that are masked. In particular, we analyse the distribution of inpainting results when masking additional regions of the image, specifically the contra-lateral structure. This allows us to elucidate where in the brain the model is drawing information from, and in particular, what is the importance of hemispherical symmetry? Our experiments interrogate a diffusion inpainting model through analysing the inpainting of subcortical brain structures based on intensity and estimated area change. We demonstrate that some structures show a strong influence of symmetry in the conditioning of the inpainting process.
Capturing Longitudinal Changes in Brain Morphology Using Temporally Parameterized Neural Displacement Fields
Apr 13, 2025Abstract:Longitudinal image registration enables studying temporal changes in brain morphology which is useful in applications where monitoring the growth or atrophy of specific structures is important. However this task is challenging due to; noise/artifacts in the data and quantifying small anatomical changes between sequential scans. We propose a novel longitudinal registration method that models structural changes using temporally parameterized neural displacement fields. Specifically, we implement an implicit neural representation (INR) using a multi-layer perceptron that serves as a continuous coordinate-based approximation of the deformation field at any time point. In effect, for any N scans of a particular subject, our model takes as input a 3D spatial coordinate location x, y, z and a corresponding temporal representation t and learns to describe the continuous morphology of structures for both observed and unobserved points in time. Furthermore, we leverage the analytic derivatives of the INR to derive a new regularization function that enforces monotonic rate of change in the trajectory of the voxels, which is shown to provide more biologically plausible patterns. We demonstrate the effectiveness of our method on 4D brain MR registration.
Unscrambling disease progression at scale: fast inference of event permutations with optimal transport
Oct 18, 2024



Abstract:Disease progression models infer group-level temporal trajectories of change in patients' features as a chronic degenerative condition plays out. They provide unique insight into disease biology and staging systems with individual-level clinical utility. Discrete models consider disease progression as a latent permutation of events, where each event corresponds to a feature becoming measurably abnormal. However, permutation inference using traditional maximum likelihood approaches becomes prohibitive due to combinatoric explosion, severely limiting model dimensionality and utility. Here we leverage ideas from optimal transport to model disease progression as a latent permutation matrix of events belonging to the Birkhoff polytope, facilitating fast inference via optimisation of the variational lower bound. This enables a factor of 1000 times faster inference than the current state of the art and, correspondingly, supports models with several orders of magnitude more features than the current state of the art can consider. Experiments demonstrate the increase in speed, accuracy and robustness to noise in simulation. Further experiments with real-world imaging data from two separate datasets, one from Alzheimer's disease patients, the other age-related macular degeneration, showcase, for the first time, pixel-level disease progression events in the brain and eye, respectively. Our method is low compute, interpretable and applicable to any progressive condition and data modality, giving it broad potential clinical utility.
Learning transition times in event sequences: the Event-Based Hidden Markov Model of disease progression
Nov 02, 2020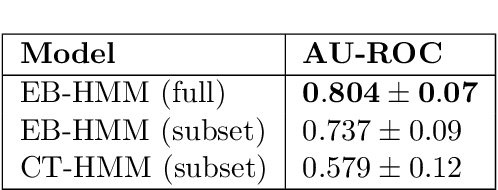
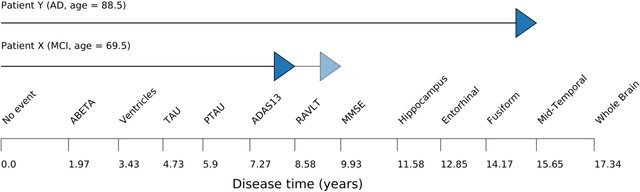
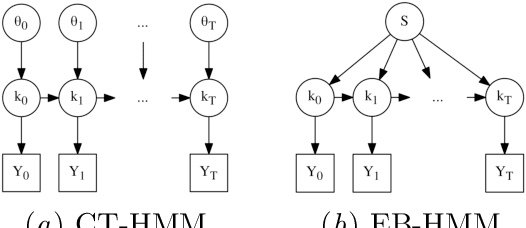
Abstract:Progressive diseases worsen over time and are characterised by monotonic change in features that track disease progression. Here we connect ideas from two formerly separate methodologies -- event-based and hidden Markov modelling -- to derive a new generative model of disease progression. Our model can uniquely infer the most likely group-level sequence and timing of events (natural history) from limited datasets. Moreover, it can infer and predict individual-level trajectories (prognosis) even when data are missing, giving it high clinical utility. Here we derive the model and provide an inference scheme based on the expectation maximisation algorithm. We use clinical, imaging and biofluid data from the Alzheimer's Disease Neuroimaging Initiative to demonstrate the validity and utility of our model. First, we train our model to uncover a new group-level sequence of feature changes in Alzheimer's disease over a period of ${\sim}17.3$ years. Next, we demonstrate that our model provides improved utility over a continuous time hidden Markov model by area under the receiver operator characteristic curve ${\sim}0.23$. Finally, we demonstrate that our model maintains predictive accuracy with up to $50\%$ missing data. These results support the clinical validity of our model and its broader utility in resource-limited medical applications.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge