Heinz Handels
Don't Mind the Gaps: Implicit Neural Representations for Resolution-Agnostic Retinal OCT Analysis
Jan 05, 2026Abstract:Routine clinical imaging of the retina using optical coherence tomography (OCT) is performed with large slice spacing, resulting in highly anisotropic images and a sparsely scanned retina. Most learning-based methods circumvent the problems arising from the anisotropy by using 2D approaches rather than performing volumetric analyses. These approaches inherently bear the risk of generating inconsistent results for neighboring B-scans. For example, 2D retinal layer segmentations can have irregular surfaces in 3D. Furthermore, the typically used convolutional neural networks are bound to the resolution of the training data, which prevents their usage for images acquired with a different imaging protocol. Implicit neural representations (INRs) have recently emerged as a tool to store voxelized data as a continuous representation. Using coordinates as input, INRs are resolution-agnostic, which allows them to be applied to anisotropic data. In this paper, we propose two frameworks that make use of this characteristic of INRs for dense 3D analyses of retinal OCT volumes. 1) We perform inter-B-scan interpolation by incorporating additional information from en-face modalities, that help retain relevant structures between B-scans. 2) We create a resolution-agnostic retinal atlas that enables general analysis without strict requirements for the data. Both methods leverage generalizable INRs, improving retinal shape representation through population-based training and allowing predictions for unseen cases. Our resolution-independent frameworks facilitate the analysis of OCT images with large B-scan distances, opening up possibilities for the volumetric evaluation of retinal structures and pathologies.
LNQ 2023 challenge: Benchmark of weakly-supervised techniques for mediastinal lymph node quantification
Aug 19, 2024



Abstract:Accurate assessment of lymph node size in 3D CT scans is crucial for cancer staging, therapeutic management, and monitoring treatment response. Existing state-of-the-art segmentation frameworks in medical imaging often rely on fully annotated datasets. However, for lymph node segmentation, these datasets are typically small due to the extensive time and expertise required to annotate the numerous lymph nodes in 3D CT scans. Weakly-supervised learning, which leverages incomplete or noisy annotations, has recently gained interest in the medical imaging community as a potential solution. Despite the variety of weakly-supervised techniques proposed, most have been validated only on private datasets or small publicly available datasets. To address this limitation, the Mediastinal Lymph Node Quantification (LNQ) challenge was organized in conjunction with the 26th International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI 2023). This challenge aimed to advance weakly-supervised segmentation methods by providing a new, partially annotated dataset and a robust evaluation framework. A total of 16 teams from 5 countries submitted predictions to the validation leaderboard, and 6 teams from 3 countries participated in the evaluation phase. The results highlighted both the potential and the current limitations of weakly-supervised approaches. On one hand, weakly-supervised approaches obtained relatively good performance with a median Dice score of $61.0\%$. On the other hand, top-ranked teams, with a median Dice score exceeding $70\%$, boosted their performance by leveraging smaller but fully annotated datasets to combine weak supervision and full supervision. This highlights both the promise of weakly-supervised methods and the ongoing need for high-quality, fully annotated data to achieve higher segmentation performance.
TSynD: Targeted Synthetic Data Generation for Enhanced Medical Image Classification
Jun 25, 2024Abstract:The usage of medical image data for the training of large-scale machine learning approaches is particularly challenging due to its scarce availability and the costly generation of data annotations, typically requiring the engagement of medical professionals. The rapid development of generative models allows towards tackling this problem by leveraging large amounts of realistic synthetically generated data for the training process. However, randomly choosing synthetic samples, might not be an optimal strategy. In this work, we investigate the targeted generation of synthetic training data, in order to improve the accuracy and robustness of image classification. Therefore, our approach aims to guide the generative model to synthesize data with high epistemic uncertainty, since large measures of epistemic uncertainty indicate underrepresented data points in the training set. During the image generation we feed images reconstructed by an auto encoder into the classifier and compute the mutual information over the class-probability distribution as a measure for uncertainty.We alter the feature space of the autoencoder through an optimization process with the objective of maximizing the classifier uncertainty on the decoded image. By training on such data we improve the performance and robustness against test time data augmentations and adversarial attacks on several classifications tasks.
LNQ Challenge 2023: Learning Mediastinal Lymph Node Segmentation with a Probabilistic Lymph Node Atlas
Jun 06, 2024



Abstract:The evaluation of lymph node metastases plays a crucial role in achieving precise cancer staging, influencing subsequent decisions regarding treatment options. Lymph node detection poses challenges due to the presence of unclear boundaries and the diverse range of sizes and morphological characteristics, making it a resource-intensive process. As part of the LNQ 2023 MICCAI challenge, we propose the use of anatomical priors as a tool to address the challenges that persist in mediastinal lymph node segmentation in combination with the partial annotation of the challenge training data. The model ensemble using all suggested modifications yields a Dice score of 0.6033 and segments 57% of the ground truth lymph nodes, compared to 27% when training on CT only. Segmentation accuracy is improved significantly by incorporating a probabilistic lymph node atlas in loss weighting and post-processing. The largest performance gains are achieved by oversampling fully annotated data to account for the partial annotation of the challenge training data, as well as adding additional data augmentation to address the high heterogeneity of the CT images and lymph node appearance. Our code is available at https://github.com/MICAI-IMI-UzL/LNQ2023.
* Accepted for publication at the Journal of Machine Learning for Biomedical Imaging (MELBA) https://melba-journal.org/2024:009
Anatomical Conditioning for Contrastive Unpaired Image-to-Image Translation of Optical Coherence Tomography Images
Apr 08, 2024Abstract:For a unified analysis of medical images from different modalities, data harmonization using image-to-image (I2I) translation is desired. We study this problem employing an optical coherence tomography (OCT) data set of Spectralis-OCT and Home-OCT images. I2I translation is challenging because the images are unpaired, and a bijective mapping does not exist due to the information discrepancy between both domains. This problem has been addressed by the Contrastive Learning for Unpaired I2I Translation (CUT) approach, but it reduces semantic consistency. To restore the semantic consistency, we support the style decoder using an additional segmentation decoder. Our approach increases the similarity between the style-translated images and the target distribution. Importantly, we improve the segmentation of biomarkers in Home-OCT images in an unsupervised domain adaptation scenario. Our data harmonization approach provides potential for the monitoring of diseases, e.g., age related macular disease, using different OCT devices.
Memory-efficient GAN-based Domain Translation of High Resolution 3D Medical Images
Oct 06, 2020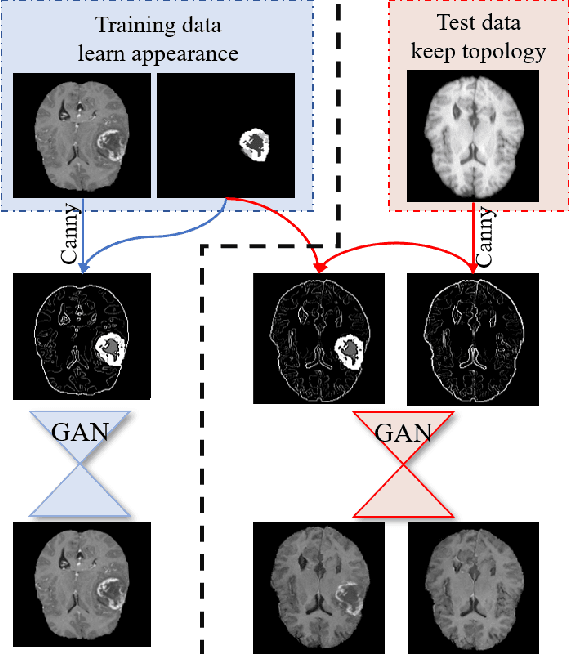
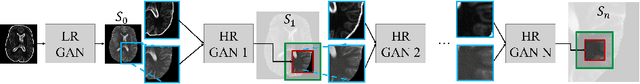

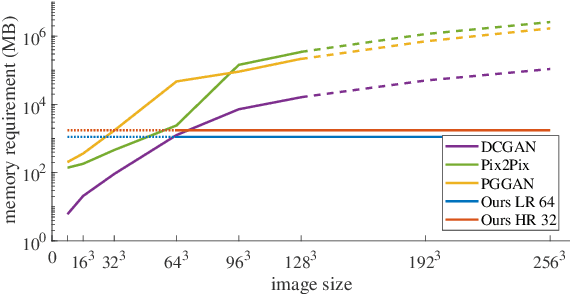
Abstract:Generative adversarial networks (GANs) are currently rarely applied on 3D medical images of large size, due to their immense computational demand. The present work proposes a multi-scale patch-based GAN approach for establishing unpaired domain translation by generating 3D medical image volumes of high resolution in a memory-efficient way. The key idea to enable memory-efficient image generation is to first generate a low-resolution version of the image followed by the generation of patches of constant sizes but successively growing resolutions. To avoid patch artifacts and incorporate global information, the patch generation is conditioned on patches from previous resolution scales. Those multi-scale GANs are trained to generate realistically looking images from image sketches in order to perform an unpaired domain translation. This allows to preserve the topology of the test data and generate the appearance of the training domain data. The evaluation of the domain translation scenarios is performed on brain MRIs of size 155x240x240 and thorax CTs of size up to 512x512x512. Compared to common patch-based approaches, the multi-resolution scheme enables better image quality and prevents patch artifacts. Also, it ensures constant GPU memory demand independent from the image size, allowing for the generation of arbitrarily large images.
Segmentation of Retinal Low-Cost Optical Coherence Tomography Images using Deep Learning
Jan 23, 2020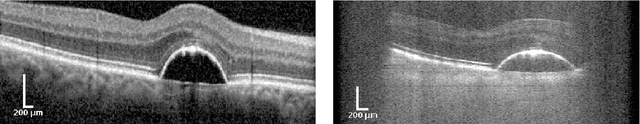
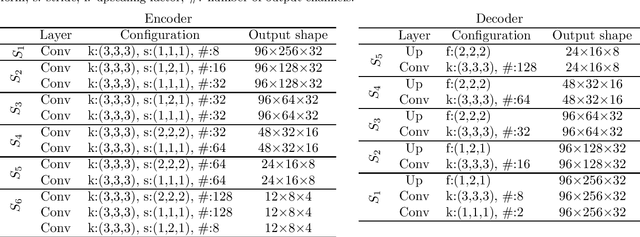
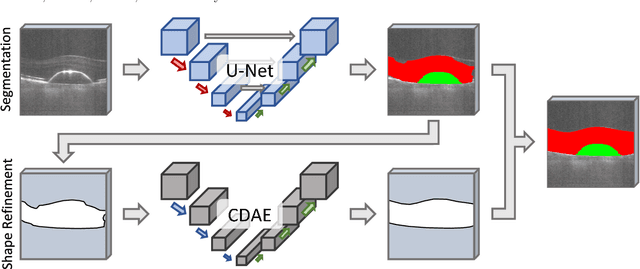
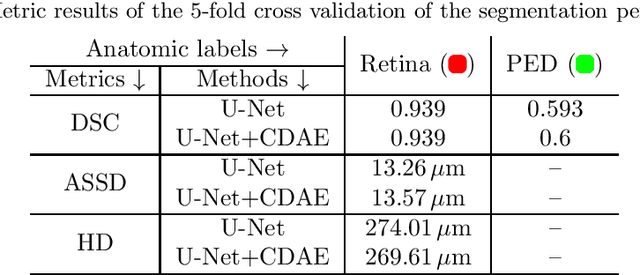
Abstract:The treatment of age-related macular degeneration (AMD) requires continuous eye exams using optical coherence tomography (OCT). The need for treatment is determined by the presence or change of disease-specific OCT-based biomarkers. Therefore, the monitoring frequency has a significant influence on the success of AMD therapy. However, the monitoring frequency of current treatment schemes is not individually adapted to the patient and therefore often insufficient. While a higher monitoring frequency would have a positive effect on the success of treatment, in practice it can only be achieved with a home monitoring solution. One of the key requirements of a home monitoring OCT system is a computer-aided diagnosis to automatically detect and quantify pathological changes using specific OCT-based biomarkers. In this paper, for the first time, retinal scans of a novel self-examination low-cost full-field OCT (SELF-OCT) are segmented using a deep learning-based approach. A convolutional neural network (CNN) is utilized to segment the total retina as well as pigment epithelial detachments (PED). It is shown that the CNN-based approach can segment the retina with high accuracy, whereas the segmentation of the PED proves to be challenging. In addition, a convolutional denoising autoencoder (CDAE) refines the CNN prediction, which has previously learned retinal shape information. It is shown that the CDAE refinement can correct segmentation errors caused by artifacts in the OCT image.
Robust GPU-based Virtual Reality Simulation of Radio Frequency Ablations for Various Needle Geometries and Locations
Jul 11, 2019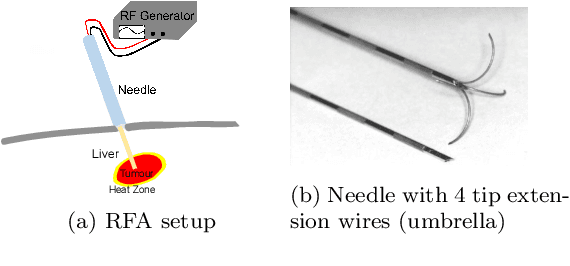

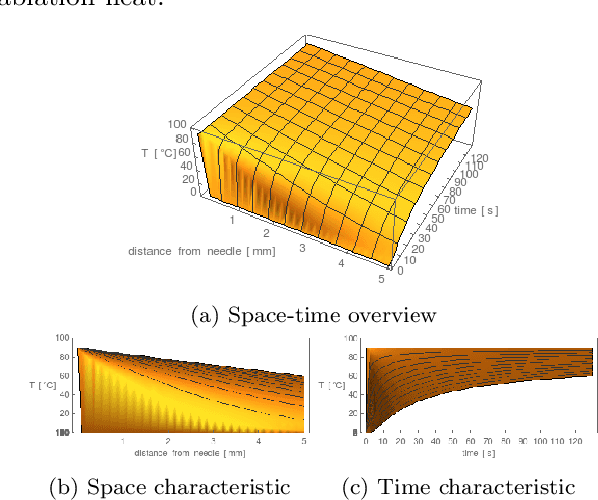

Abstract:Purpose: Radio-frequency ablations play an important role in the therapy of malignant liver lesions. The navigation of a needle to the lesion poses a challenge for both the trainees and intervening physicians. Methods: This publication presents a new GPU-based, accurate method for the simulation of radio-frequency ablations for lesions at the needle tip in general and for an existing visuo-haptic 4D VR simulator. The method is implemented real-time capable with Nvidia CUDA. Results: It performs better than a literature method concerning the theoretical characteristic of monotonic convergence of the bioheat PDE and a in vitro gold standard with significant improvements (p < 0.05) in terms of Pearson correlations. It shows no failure modes or theoretically inconsistent individual simulation results after the initial phase of 10 seconds. On the Nvidia 1080 Ti GPU it achieves a very high frame rendering performance of >480 Hz. Conclusion: Our method provides a more robust and safer real-time ablation planning and intraoperative guidance technique, especially avoiding the over-estimation of the ablated tissue death zone, which is risky for the patient in terms of tumor recurrence. Future in vitro measurements and optimization shall further improve the conservative estimate.
* 18 pages, 14 figures, 1 table, 2 algorithms, 2 movies
Multi-scale GANs for Memory-efficient Generation of High Resolution Medical Images
Jul 08, 2019



Abstract:Currently generative adversarial networks (GANs) are rarely applied to medical images of large sizes, especially 3D volumes, due to their large computational demand. We propose a novel multi-scale patch-based GAN approach to generate large high resolution 2D and 3D images. Our key idea is to first learn a low-resolution version of the image and then generate patches of successively growing resolutions conditioned on previous scales. In a domain translation use-case scenario, 3D thorax CTs of size 512x512x512 and thorax X-rays of size 2048x2048 are generated and we show that, due to the constant GPU memory demand of our method, arbitrarily large images of high resolution can be generated. Moreover, compared to common patch-based approaches, our multi-resolution scheme enables better image quality and prevents patch artifacts.
Estimation of Large Motion in Lung CT by Integrating Regularized Keypoint Correspondences into Dense Deformable Registration
Jul 02, 2018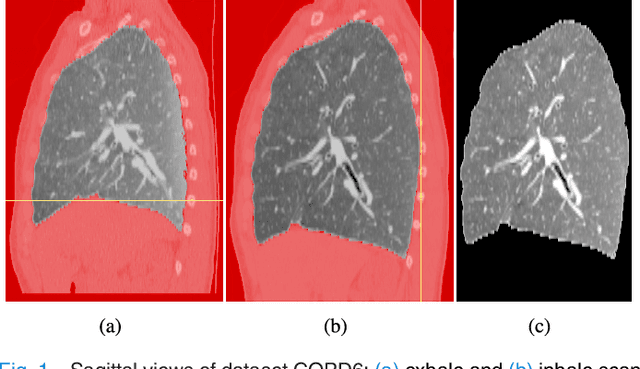
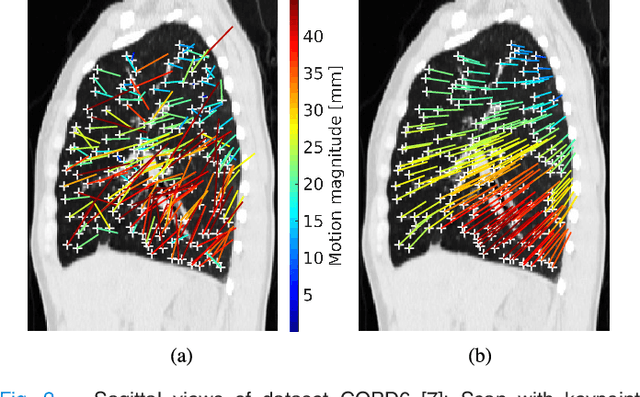
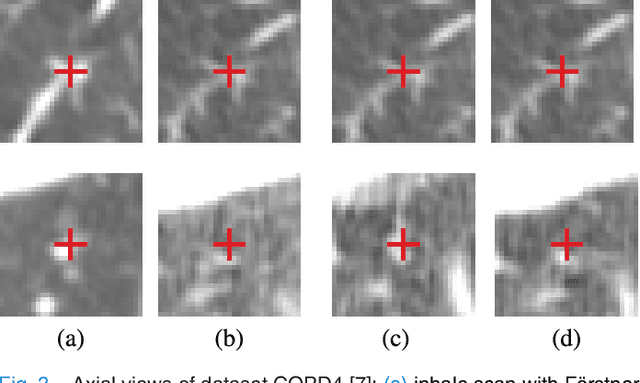
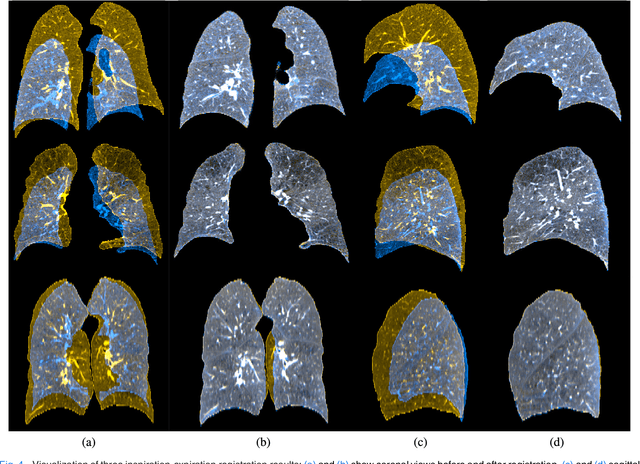
Abstract:We present a novel algorithm for the registration of pulmonary CT scans. Our method is designed for large respiratory motion by integrating sparse keypoint correspondences into a dense continuous optimization framework. The detection of keypoint correspondences enables robustness against large deformations by jointly optimizing over a large number of potential discrete displacements, whereas the dense continuous registration achieves subvoxel alignment with smooth transformations. Both steps are driven by the same normalized gradient fields data term. We employ curvature regularization and a volume change control mechanism to prevent foldings of the deformation grid and restrict the determinant of the Jacobian to physiologically meaningful values. Keypoint correspondences are integrated into the dense registration by a quadratic penalty with adaptively determined weight. Using a parallel matrix-free derivative calculation scheme, a runtime of about 5 min was realized on a standard PC. The proposed algorithm ranks first in the EMPIRE10 challenge on pulmonary image registration. Moreover, it achieves an average landmark distance of 0.82 mm on the DIR-Lab COPD database, thereby improving upon the state of the art in accuracy by 15%. Our algorithm is the first to reach the inter-observer variability in landmark annotation on this dataset.
* 12 pages, 7 figures, \c{opyright} 2017 IEEE. Personal use is permitted, but republication/redistribution requires IEEE permission
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge