Timo Kepp
Don't Mind the Gaps: Implicit Neural Representations for Resolution-Agnostic Retinal OCT Analysis
Jan 05, 2026Abstract:Routine clinical imaging of the retina using optical coherence tomography (OCT) is performed with large slice spacing, resulting in highly anisotropic images and a sparsely scanned retina. Most learning-based methods circumvent the problems arising from the anisotropy by using 2D approaches rather than performing volumetric analyses. These approaches inherently bear the risk of generating inconsistent results for neighboring B-scans. For example, 2D retinal layer segmentations can have irregular surfaces in 3D. Furthermore, the typically used convolutional neural networks are bound to the resolution of the training data, which prevents their usage for images acquired with a different imaging protocol. Implicit neural representations (INRs) have recently emerged as a tool to store voxelized data as a continuous representation. Using coordinates as input, INRs are resolution-agnostic, which allows them to be applied to anisotropic data. In this paper, we propose two frameworks that make use of this characteristic of INRs for dense 3D analyses of retinal OCT volumes. 1) We perform inter-B-scan interpolation by incorporating additional information from en-face modalities, that help retain relevant structures between B-scans. 2) We create a resolution-agnostic retinal atlas that enables general analysis without strict requirements for the data. Both methods leverage generalizable INRs, improving retinal shape representation through population-based training and allowing predictions for unseen cases. Our resolution-independent frameworks facilitate the analysis of OCT images with large B-scan distances, opening up possibilities for the volumetric evaluation of retinal structures and pathologies.
LNQ Challenge 2023: Learning Mediastinal Lymph Node Segmentation with a Probabilistic Lymph Node Atlas
Jun 06, 2024



Abstract:The evaluation of lymph node metastases plays a crucial role in achieving precise cancer staging, influencing subsequent decisions regarding treatment options. Lymph node detection poses challenges due to the presence of unclear boundaries and the diverse range of sizes and morphological characteristics, making it a resource-intensive process. As part of the LNQ 2023 MICCAI challenge, we propose the use of anatomical priors as a tool to address the challenges that persist in mediastinal lymph node segmentation in combination with the partial annotation of the challenge training data. The model ensemble using all suggested modifications yields a Dice score of 0.6033 and segments 57% of the ground truth lymph nodes, compared to 27% when training on CT only. Segmentation accuracy is improved significantly by incorporating a probabilistic lymph node atlas in loss weighting and post-processing. The largest performance gains are achieved by oversampling fully annotated data to account for the partial annotation of the challenge training data, as well as adding additional data augmentation to address the high heterogeneity of the CT images and lymph node appearance. Our code is available at https://github.com/MICAI-IMI-UzL/LNQ2023.
* Accepted for publication at the Journal of Machine Learning for Biomedical Imaging (MELBA) https://melba-journal.org/2024:009
Anatomical Conditioning for Contrastive Unpaired Image-to-Image Translation of Optical Coherence Tomography Images
Apr 08, 2024Abstract:For a unified analysis of medical images from different modalities, data harmonization using image-to-image (I2I) translation is desired. We study this problem employing an optical coherence tomography (OCT) data set of Spectralis-OCT and Home-OCT images. I2I translation is challenging because the images are unpaired, and a bijective mapping does not exist due to the information discrepancy between both domains. This problem has been addressed by the Contrastive Learning for Unpaired I2I Translation (CUT) approach, but it reduces semantic consistency. To restore the semantic consistency, we support the style decoder using an additional segmentation decoder. Our approach increases the similarity between the style-translated images and the target distribution. Importantly, we improve the segmentation of biomarkers in Home-OCT images in an unsupervised domain adaptation scenario. Our data harmonization approach provides potential for the monitoring of diseases, e.g., age related macular disease, using different OCT devices.
Segmentation of Retinal Low-Cost Optical Coherence Tomography Images using Deep Learning
Jan 23, 2020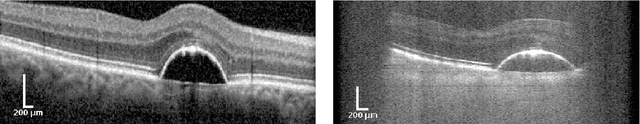
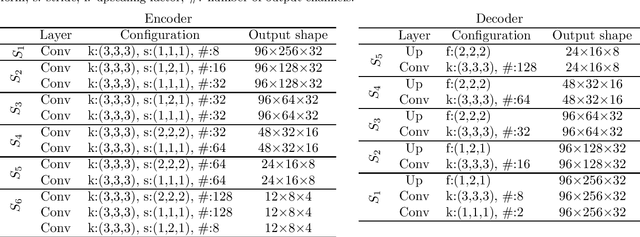
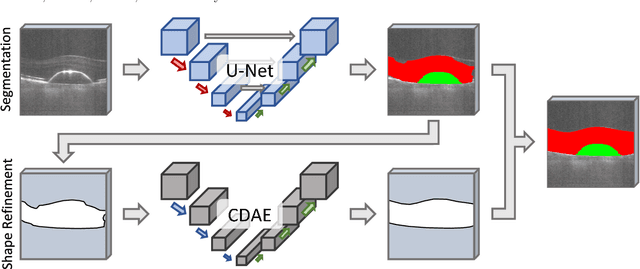
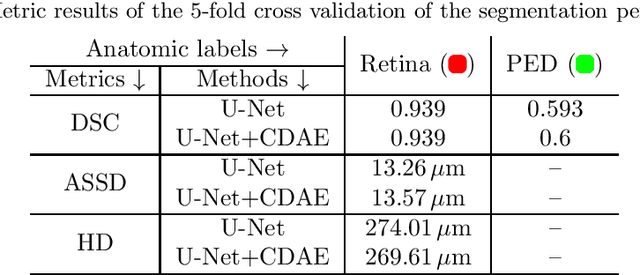
Abstract:The treatment of age-related macular degeneration (AMD) requires continuous eye exams using optical coherence tomography (OCT). The need for treatment is determined by the presence or change of disease-specific OCT-based biomarkers. Therefore, the monitoring frequency has a significant influence on the success of AMD therapy. However, the monitoring frequency of current treatment schemes is not individually adapted to the patient and therefore often insufficient. While a higher monitoring frequency would have a positive effect on the success of treatment, in practice it can only be achieved with a home monitoring solution. One of the key requirements of a home monitoring OCT system is a computer-aided diagnosis to automatically detect and quantify pathological changes using specific OCT-based biomarkers. In this paper, for the first time, retinal scans of a novel self-examination low-cost full-field OCT (SELF-OCT) are segmented using a deep learning-based approach. A convolutional neural network (CNN) is utilized to segment the total retina as well as pigment epithelial detachments (PED). It is shown that the CNN-based approach can segment the retina with high accuracy, whereas the segmentation of the PED proves to be challenging. In addition, a convolutional denoising autoencoder (CDAE) refines the CNN prediction, which has previously learned retinal shape information. It is shown that the CDAE refinement can correct segmentation errors caused by artifacts in the OCT image.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge