Hristina Uzunova
TSynD: Targeted Synthetic Data Generation for Enhanced Medical Image Classification
Jun 25, 2024Abstract:The usage of medical image data for the training of large-scale machine learning approaches is particularly challenging due to its scarce availability and the costly generation of data annotations, typically requiring the engagement of medical professionals. The rapid development of generative models allows towards tackling this problem by leveraging large amounts of realistic synthetically generated data for the training process. However, randomly choosing synthetic samples, might not be an optimal strategy. In this work, we investigate the targeted generation of synthetic training data, in order to improve the accuracy and robustness of image classification. Therefore, our approach aims to guide the generative model to synthesize data with high epistemic uncertainty, since large measures of epistemic uncertainty indicate underrepresented data points in the training set. During the image generation we feed images reconstructed by an auto encoder into the classifier and compute the mutual information over the class-probability distribution as a measure for uncertainty.We alter the feature space of the autoencoder through an optimization process with the objective of maximizing the classifier uncertainty on the decoded image. By training on such data we improve the performance and robustness against test time data augmentations and adversarial attacks on several classifications tasks.
Anatomical Conditioning for Contrastive Unpaired Image-to-Image Translation of Optical Coherence Tomography Images
Apr 08, 2024Abstract:For a unified analysis of medical images from different modalities, data harmonization using image-to-image (I2I) translation is desired. We study this problem employing an optical coherence tomography (OCT) data set of Spectralis-OCT and Home-OCT images. I2I translation is challenging because the images are unpaired, and a bijective mapping does not exist due to the information discrepancy between both domains. This problem has been addressed by the Contrastive Learning for Unpaired I2I Translation (CUT) approach, but it reduces semantic consistency. To restore the semantic consistency, we support the style decoder using an additional segmentation decoder. Our approach increases the similarity between the style-translated images and the target distribution. Importantly, we improve the segmentation of biomarkers in Home-OCT images in an unsupervised domain adaptation scenario. Our data harmonization approach provides potential for the monitoring of diseases, e.g., age related macular disease, using different OCT devices.
Memory-efficient GAN-based Domain Translation of High Resolution 3D Medical Images
Oct 06, 2020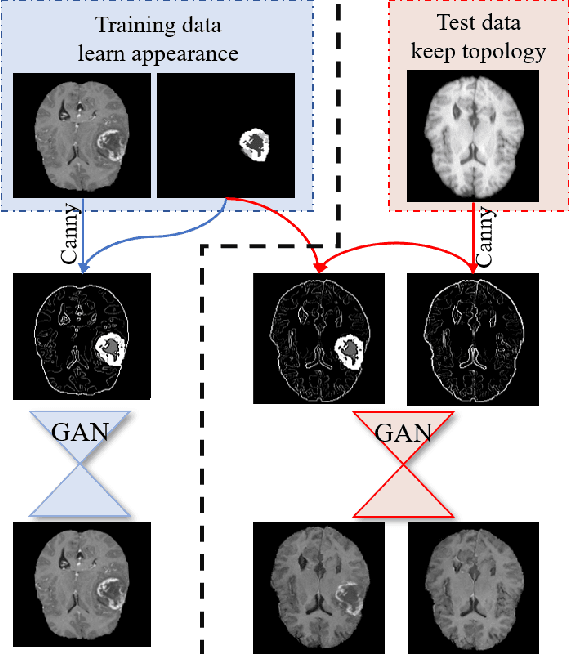
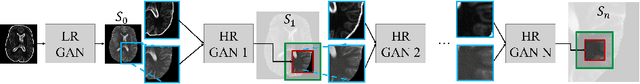

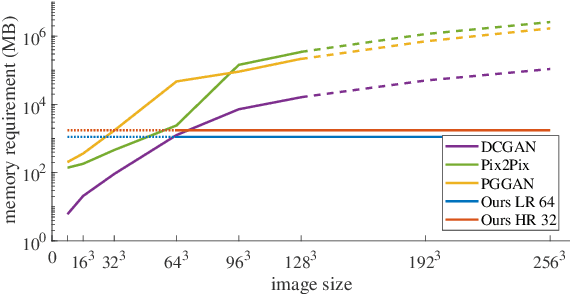
Abstract:Generative adversarial networks (GANs) are currently rarely applied on 3D medical images of large size, due to their immense computational demand. The present work proposes a multi-scale patch-based GAN approach for establishing unpaired domain translation by generating 3D medical image volumes of high resolution in a memory-efficient way. The key idea to enable memory-efficient image generation is to first generate a low-resolution version of the image followed by the generation of patches of constant sizes but successively growing resolutions. To avoid patch artifacts and incorporate global information, the patch generation is conditioned on patches from previous resolution scales. Those multi-scale GANs are trained to generate realistically looking images from image sketches in order to perform an unpaired domain translation. This allows to preserve the topology of the test data and generate the appearance of the training domain data. The evaluation of the domain translation scenarios is performed on brain MRIs of size 155x240x240 and thorax CTs of size up to 512x512x512. Compared to common patch-based approaches, the multi-resolution scheme enables better image quality and prevents patch artifacts. Also, it ensures constant GPU memory demand independent from the image size, allowing for the generation of arbitrarily large images.
Multi-scale GANs for Memory-efficient Generation of High Resolution Medical Images
Jul 08, 2019



Abstract:Currently generative adversarial networks (GANs) are rarely applied to medical images of large sizes, especially 3D volumes, due to their large computational demand. We propose a novel multi-scale patch-based GAN approach to generate large high resolution 2D and 3D images. Our key idea is to first learn a low-resolution version of the image and then generate patches of successively growing resolutions conditioned on previous scales. In a domain translation use-case scenario, 3D thorax CTs of size 512x512x512 and thorax X-rays of size 2048x2048 are generated and we show that, due to the constant GPU memory demand of our method, arbitrarily large images of high resolution can be generated. Moreover, compared to common patch-based approaches, our multi-resolution scheme enables better image quality and prevents patch artifacts.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge