Ivan Olier
Conditional Diffusion-Flow models for generating 3D cosmic density fields: applications to f(R) cosmologies
Feb 24, 2025Abstract:Next-generation galaxy surveys promise unprecedented precision in testing gravity at cosmological scales. However, realising this potential requires accurately modelling the non-linear cosmic web. We address this challenge by exploring conditional generative modelling to create 3D dark matter density fields via score-based (diffusion) and flow-based methods. Our results demonstrate the power of diffusion models to accurately reproduce the matter power spectra and bispectra, even for unseen configurations. They also offer a significant speed-up with slightly reduced accuracy, when flow-based reconstructing the probability distribution function, but they struggle with higher-order statistics. To improve conditional generation, we introduce a novel multi-output model to develop feature representations of the cosmological parameters. Our findings offer a powerful tool for exploring deviations from standard gravity, combining high precision with reduced computational cost, thus paving the way for more comprehensive and efficient cosmological analyses
The Partial Response Network
Aug 16, 2019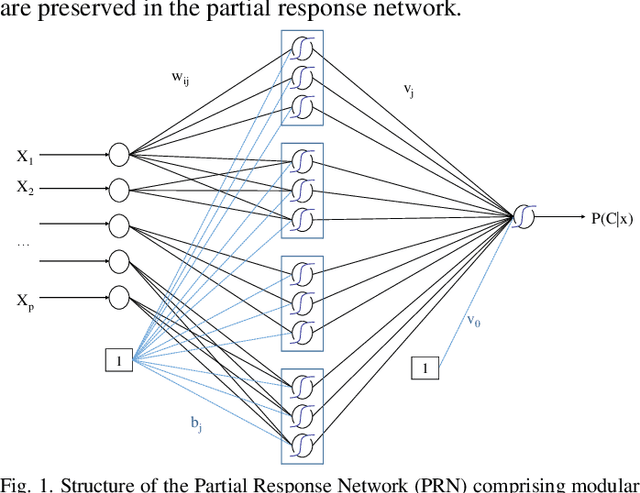
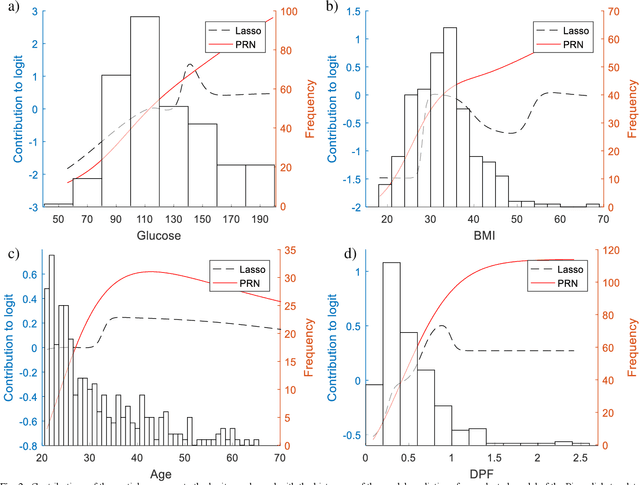
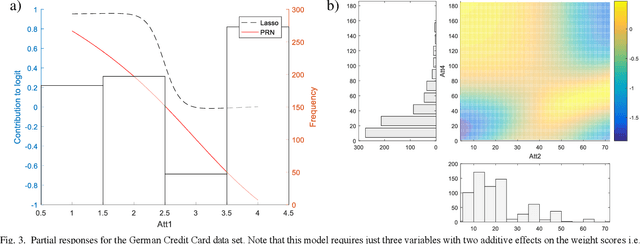
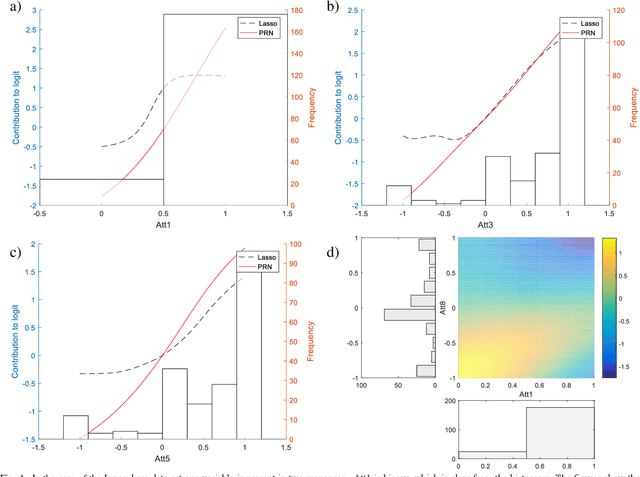
Abstract:We propose a method to open the black box of the Multi-Layer Perceptron by inferring from it a simpler and generally more accurate general additive model. The resulting model comprises non-linear univariate and bivariate partial responses derived from the original Multi-Layer Perceptron. The responses are combined using the Lasso and further optimised within a modular structure. The approach is generic and provides a constructive framework to simplify and explain the Multi-Layer Perceptron for any data set, opening the door for validation against prior knowledge. Experimental results on benchmarking datasets indicate that the partial responses are intuitive to interpret and the Area Under the Curve is competitive with Gradient Boosting, Support Vector Machines and Random Forests. The performance improvement compared with a fully connected Multi-Layer Perceptron is attributed to reduced confounding in the second stage of optimisation of the weights. The main limitation of the method is that it explicitly models only up to pairwise interactions. For many practical applications this will be optimal, but where that is not the case then this will be indicated by the performance difference compared to the original model. The streamlined model simultaneously interprets and optimises this frequently used flexible model.
Transformative Machine Learning
Nov 08, 2018



Abstract:The key to success in machine learning (ML) is the use of effective data representations. Traditionally, data representations were hand-crafted. Recently it has been demonstrated that, given sufficient data, deep neural networks can learn effective implicit representations from simple input representations. However, for most scientific problems, the use of deep learning is not appropriate as the amount of available data is limited, and/or the output models must be explainable. Nevertheless, many scientific problems do have significant amounts of data available on related tasks, which makes them amenable to multi-task learning, i.e. learning many related problems simultaneously. Here we propose a novel and general representation learning approach for multi-task learning that works successfully with small amounts of data. The fundamental new idea is to transform an input intrinsic data representation (i.e., handcrafted features), to an extrinsic representation based on what a pre-trained set of models predict about the examples. This transformation has the dual advantages of producing significantly more accurate predictions, and providing explainable models. To demonstrate the utility of this transformative learning approach, we have applied it to three real-world scientific problems: drug-design (quantitative structure activity relationship learning), predicting human gene expression (across different tissue types and drug treatments), and meta-learning for machine learning (predicting which machine learning methods work best for a given problem). In all three problems, transformative machine learning significantly outperforms the best intrinsic representation.
Meta-QSAR: a large-scale application of meta-learning to drug design and discovery
Sep 12, 2017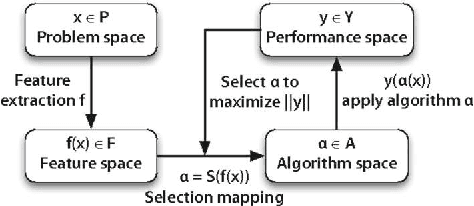
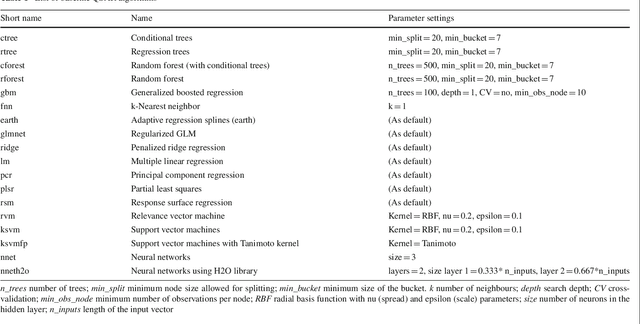

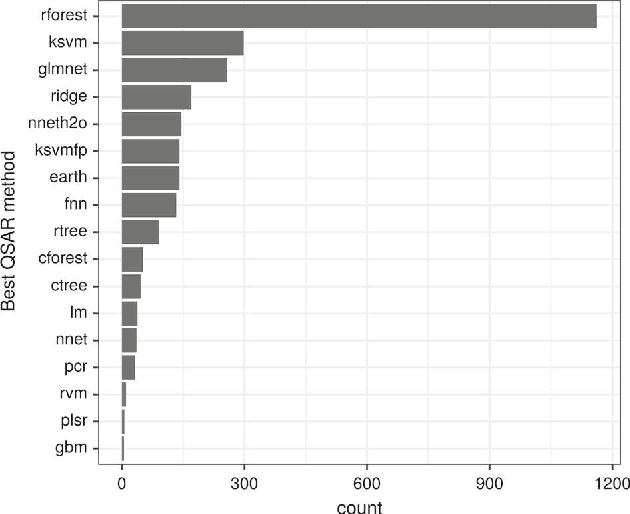
Abstract:We investigate the learning of quantitative structure activity relationships (QSARs) as a case-study of meta-learning. This application area is of the highest societal importance, as it is a key step in the development of new medicines. The standard QSAR learning problem is: given a target (usually a protein) and a set of chemical compounds (small molecules) with associated bioactivities (e.g. inhibition of the target), learn a predictive mapping from molecular representation to activity. Although almost every type of machine learning method has been applied to QSAR learning there is no agreed single best way of learning QSARs, and therefore the problem area is well-suited to meta-learning. We first carried out the most comprehensive ever comparison of machine learning methods for QSAR learning: 18 regression methods, 6 molecular representations, applied to more than 2,700 QSAR problems. (These results have been made publicly available on OpenML and represent a valuable resource for testing novel meta-learning methods.) We then investigated the utility of algorithm selection for QSAR problems. We found that this meta-learning approach outperformed the best individual QSAR learning method (random forests using a molecular fingerprint representation) by up to 13%, on average. We conclude that meta-learning outperforms base-learning methods for QSAR learning, and as this investigation is one of the most extensive ever comparisons of base and meta-learning methods ever made, it provides evidence for the general effectiveness of meta-learning over base-learning.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge