Israt Jahan
Lemon and Orange Disease Classification using CNN-Extracted Features and Machine Learning Classifier
Aug 26, 2024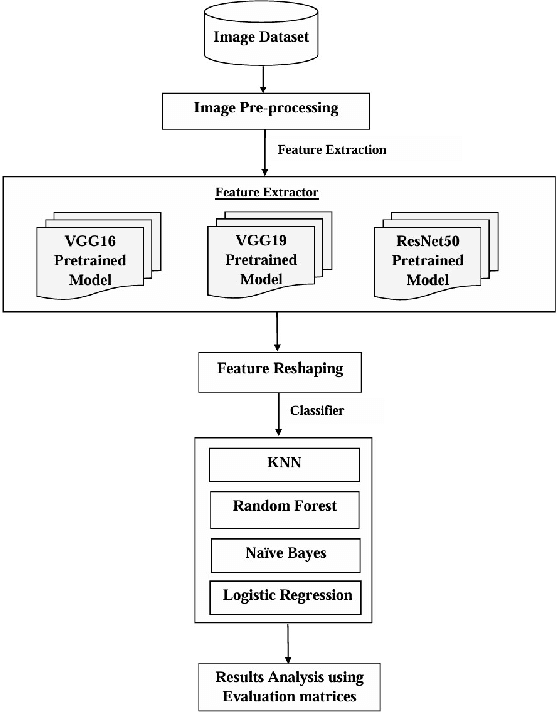


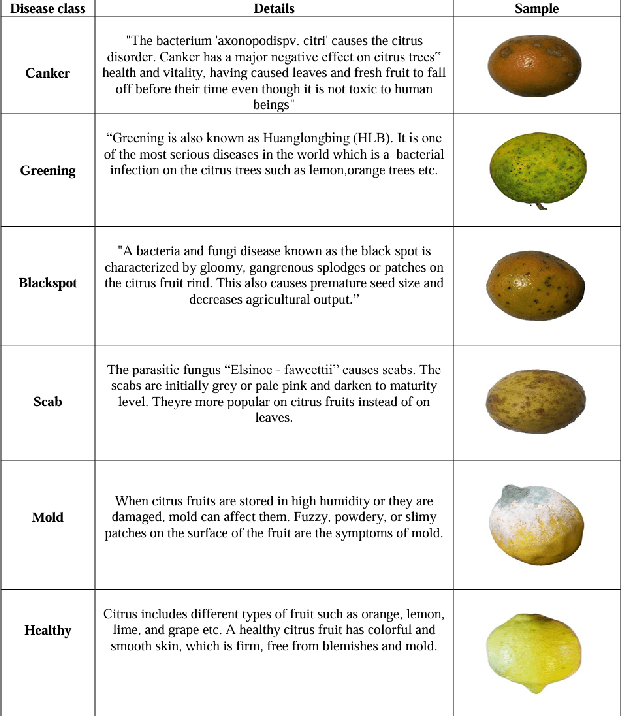
Abstract:Lemons and oranges, both are the most economically significant citrus fruits globally. The production of lemons and oranges is severely affected due to diseases in its growth stages. Fruit quality has degraded due to the presence of flaws. Thus, it is necessary to diagnose the disease accurately so that we can avoid major loss of lemons and oranges. To improve citrus farming, we proposed a disease classification approach for lemons and oranges. This approach would enable early disease detection and intervention, reduce yield losses, and optimize resource allocation. For the initial modeling of disease classification, the research uses innovative deep learning architectures such as VGG16, VGG19 and ResNet50. In addition, for achieving better accuracy, the basic machine learning algorithms used for classification problems include Random Forest, Naive Bayes, K-Nearest Neighbors (KNN) and Logistic Regression. The lemon and orange fruits diseases are classified more accurately (95.0% for lemon and 99.69% for orange) by the model. The model's base features were extracted from the ResNet50 pre-trained model and the diseases are classified by the Logistic Regression which beats the performance given by VGG16 and VGG19 for other classifiers. Experimental outcomes show that the proposed model also outperforms existing models in which most of them classified the diseases using the Softmax classifier without using any individual classifiers.
A Systematic Survey and Critical Review on Evaluating Large Language Models: Challenges, Limitations, and Recommendations
Jul 04, 2024



Abstract:Large Language Models (LLMs) have recently gained significant attention due to their remarkable capabilities in performing diverse tasks across various domains. However, a thorough evaluation of these models is crucial before deploying them in real-world applications to ensure they produce reliable performance. Despite the well-established importance of evaluating LLMs in the community, the complexity of the evaluation process has led to varied evaluation setups, causing inconsistencies in findings and interpretations. To address this, we systematically review the primary challenges and limitations causing these inconsistencies and unreliable evaluations in various steps of LLM evaluation. Based on our critical review, we present our perspectives and recommendations to ensure LLM evaluations are reproducible, reliable, and robust.
A Comprehensive Evaluation of Large Language Models on Benchmark Biomedical Text Processing Tasks
Oct 10, 2023



Abstract:Recently, Large Language Models (LLM) have demonstrated impressive capability to solve a wide range of tasks. However, despite their success across various tasks, no prior work has investigated their capability in the biomedical domain yet. To this end, this paper aims to evaluate the performance of LLMs on benchmark biomedical tasks. For this purpose, we conduct a comprehensive evaluation of 4 popular LLMs in 6 diverse biomedical tasks across 26 datasets. To the best of our knowledge, this is the first work that conducts an extensive evaluation and comparison of various LLMs in the biomedical domain. Interestingly, we find based on our evaluation that in biomedical datasets that have smaller training sets, zero-shot LLMs even outperform the current state-of-the-art fine-tuned biomedical models. This suggests that pretraining on large text corpora makes LLMs quite specialized even in the biomedical domain. We also find that not a single LLM can outperform other LLMs in all tasks, with the performance of different LLMs may vary depending on the task. While their performance is still quite poor in comparison to the biomedical models that were fine-tuned on large training sets, our findings demonstrate that LLMs have the potential to be a valuable tool for various biomedical tasks that lack large annotated data.
Evaluation of ChatGPT on Biomedical Tasks: A Zero-Shot Comparison with Fine-Tuned Generative Transformers
Jun 07, 2023



Abstract:ChatGPT is a large language model developed by OpenAI. Despite its impressive performance across various tasks, no prior work has investigated its capability in the biomedical domain yet. To this end, this paper aims to evaluate the performance of ChatGPT on various benchmark biomedical tasks, such as relation extraction, document classification, question answering, and summarization. To the best of our knowledge, this is the first work that conducts an extensive evaluation of ChatGPT in the biomedical domain. Interestingly, we find based on our evaluation that in biomedical datasets that have smaller training sets, zero-shot ChatGPT even outperforms the state-of-the-art fine-tuned generative transformer models, such as BioGPT and BioBART. This suggests that ChatGPT's pre-training on large text corpora makes it quite specialized even in the biomedical domain. Our findings demonstrate that ChatGPT has the potential to be a valuable tool for various tasks in the biomedical domain that lack large annotated data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge