Indrajit Saha
COBRA: Contextual Bandit Algorithm for Ensuring Truthful Strategic Agents
May 29, 2025Abstract:This paper considers a contextual bandit problem involving multiple agents, where a learner sequentially observes the contexts and the agent's reported arms, and then selects the arm that maximizes the system's overall reward. Existing work in contextual bandits assumes that agents truthfully report their arms, which is unrealistic in many real-life applications. For instance, consider an online platform with multiple sellers; some sellers may misrepresent product quality to gain an advantage, such as having the platform preferentially recommend their products to online users. To address this challenge, we propose an algorithm, COBRA, for contextual bandit problems involving strategic agents that disincentivize their strategic behavior without using any monetary incentives, while having incentive compatibility and a sub-linear regret guarantee. Our experimental results also validate the different performance aspects of our proposed algorithm.
A Review on the Applications of Transformer-based language models for Nucleotide Sequence Analysis
Dec 10, 2024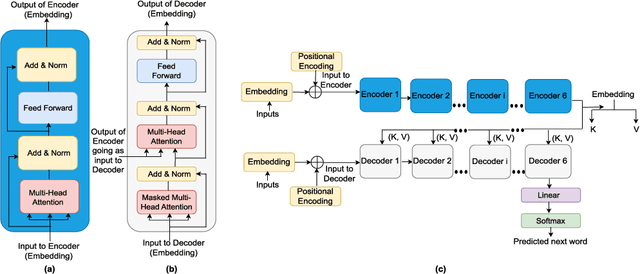

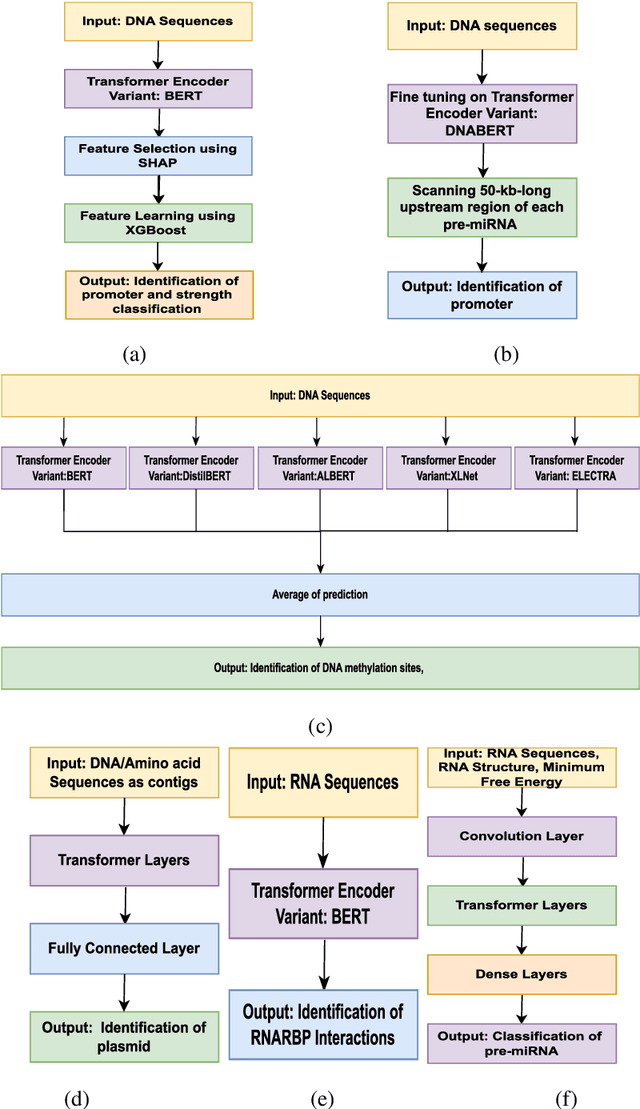
Abstract:In recent times, Transformer-based language models are making quite an impact in the field of natural language processing. As relevant parallels can be drawn between biological sequences and natural languages, the models used in NLP can be easily extended and adapted for various applications in bioinformatics. In this regard, this paper introduces the major developments of Transformer-based models in the recent past in the context of nucleotide sequences. We have reviewed and analysed a large number of application-based papers on this subject, giving evidence of the main characterizing features and to different approaches that may be adopted to customize such powerful computational machines. We have also provided a structured description of the functioning of Transformers, that may enable even first time users to grab the essence of such complex architectures. We believe this review will help the scientific community in understanding the various applications of Transformer-based language models to nucleotide sequences. This work will motivate the readers to build on these methodologies to tackle also various other problems in the field of bioinformatics.
Online Fair Division with Contextual Bandits
Aug 23, 2024Abstract:This paper considers a novel online fair division problem involving multiple agents in which a learner observes an indivisible item that has to be irrevocably allocated to one of the agents while satisfying a fairness and efficiency constraint. Existing algorithms assume a small number of items with a sufficiently large number of copies, which ensures a good utility estimation for all item-agent pairs. However, such an assumption may not hold in many real-life applications, e.g., an online platform that has a large number of users (items) who only use the platform's service providers (agents) a few times (a few copies of items), which makes it difficult to estimate the utility for all item-agent pairs. To overcome this challenge, we model the online fair division problem using contextual bandits, assuming the utility is an unknown function of the item-agent features. We then propose algorithms for online fair division with sub-linear regret guarantees. Our experimental results also verify the different performance aspects of the proposed algorithms.
Predicting Transcription Factor Binding Sites using Transformer based Capsule Network
Oct 23, 2023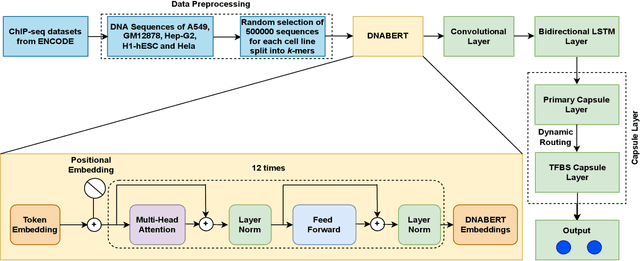
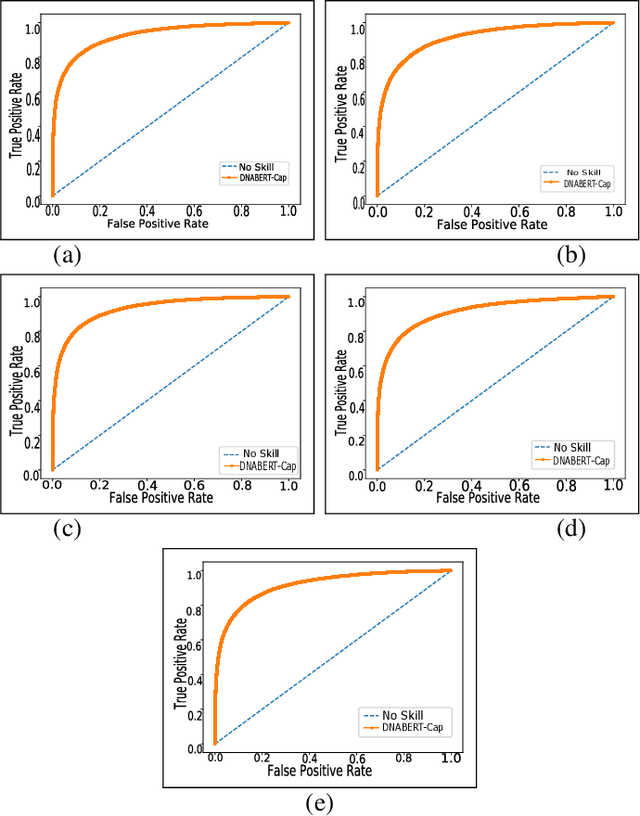
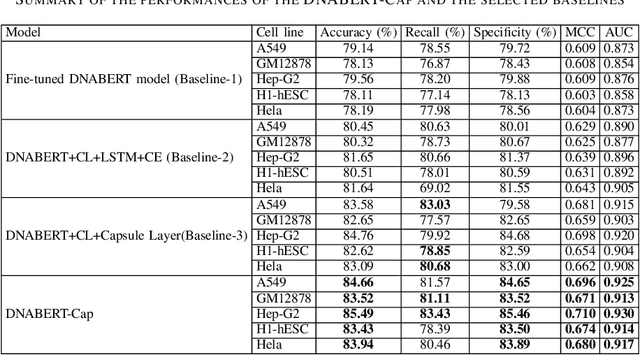
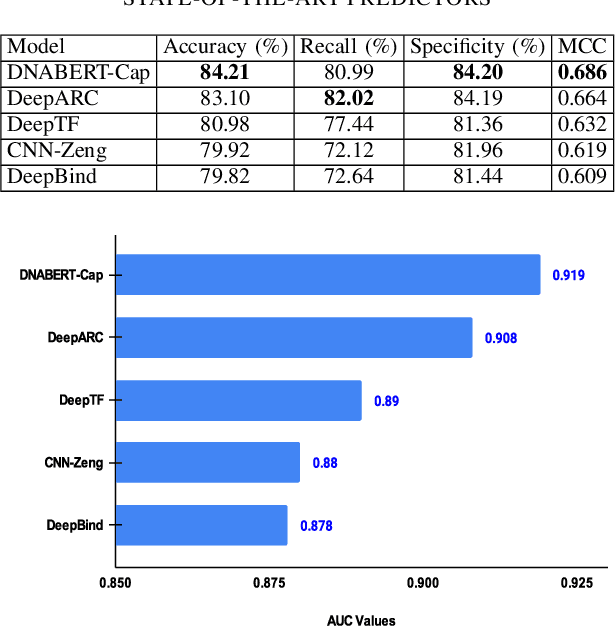
Abstract:Prediction of binding sites for transcription factors is important to understand how they regulate gene expression and how this regulation can be modulated for therapeutic purposes. Although in the past few years there are significant works addressing this issue, there is still space for improvement. In this regard, a transformer based capsule network viz. DNABERT-Cap is proposed in this work to predict transcription factor binding sites mining ChIP-seq datasets. DNABERT-Cap is a bidirectional encoder pre-trained with large number of genomic DNA sequences, empowered with a capsule layer responsible for the final prediction. The proposed model builds a predictor for transcription factor binding sites using the joint optimisation of features encompassing both bidirectional encoder and capsule layer, along with convolutional and bidirectional long-short term memory layers. To evaluate the efficiency of the proposed approach, we use a benchmark ChIP-seq datasets of five cell lines viz. A549, GM12878, Hep-G2, H1-hESC and Hela, available in the ENCODE repository. The results show that the average area under the receiver operating characteristic curve score exceeds 0.91 for all such five cell lines. DNABERT-Cap is also compared with existing state-of-the-art deep learning based predictors viz. DeepARC, DeepTF, CNN-Zeng and DeepBind, and is seen to outperform them.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge