Giovanni Felici
A Review on the Applications of Transformer-based language models for Nucleotide Sequence Analysis
Dec 10, 2024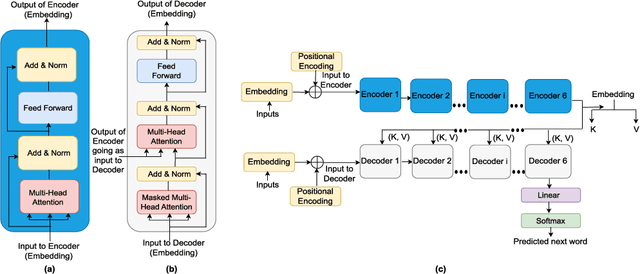

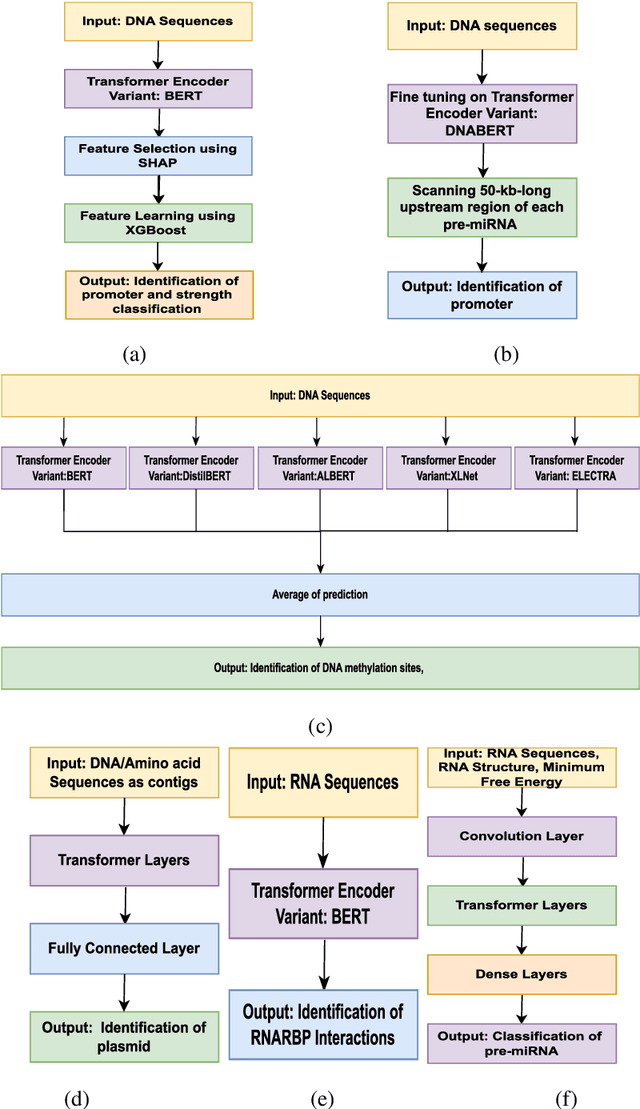
Abstract:In recent times, Transformer-based language models are making quite an impact in the field of natural language processing. As relevant parallels can be drawn between biological sequences and natural languages, the models used in NLP can be easily extended and adapted for various applications in bioinformatics. In this regard, this paper introduces the major developments of Transformer-based models in the recent past in the context of nucleotide sequences. We have reviewed and analysed a large number of application-based papers on this subject, giving evidence of the main characterizing features and to different approaches that may be adopted to customize such powerful computational machines. We have also provided a structured description of the functioning of Transformers, that may enable even first time users to grab the essence of such complex architectures. We believe this review will help the scientific community in understanding the various applications of Transformer-based language models to nucleotide sequences. This work will motivate the readers to build on these methodologies to tackle also various other problems in the field of bioinformatics.
Multi-task learning of convex combinations of forecasting models
Oct 31, 2023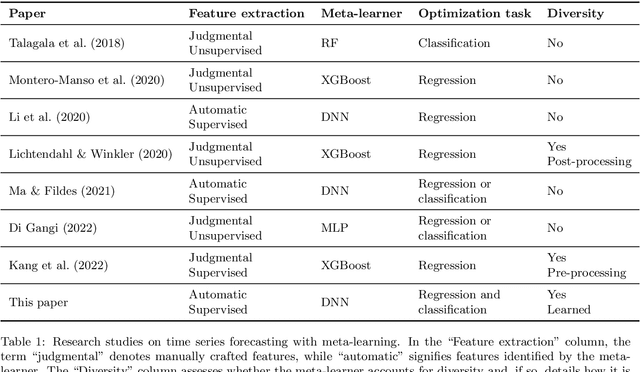
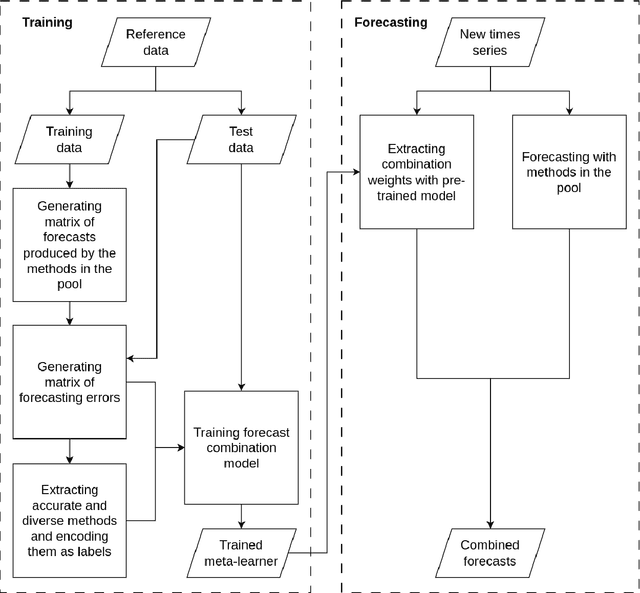
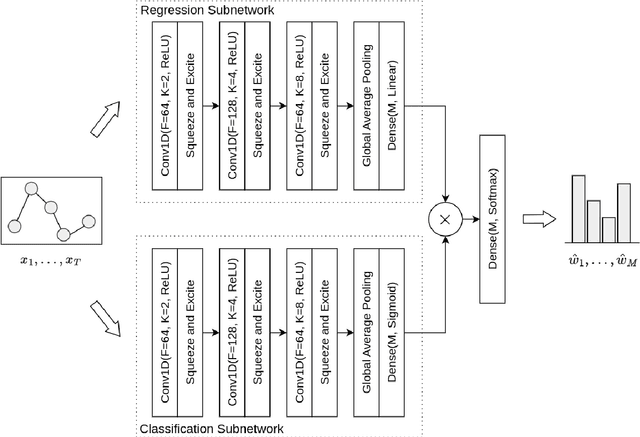
Abstract:Forecast combination involves using multiple forecasts to create a single, more accurate prediction. Recently, feature-based forecasting has been employed to either select the most appropriate forecasting models or to learn the weights of their convex combination. In this paper, we present a multi-task learning methodology that simultaneously addresses both problems. This approach is implemented through a deep neural network with two branches: the regression branch, which learns the weights of various forecasting methods by minimizing the error of combined forecasts, and the classification branch, which selects forecasting methods with an emphasis on their diversity. To generate training labels for the classification task, we introduce an optimization-driven approach that identifies the most appropriate methods for a given time series. The proposed approach elicits the essential role of diversity in feature-based forecasting and highlights the interplay between model combination and model selection when learning forecasting ensembles. Experimental results on a large set of series from the M4 competition dataset show that our proposal enhances point forecast accuracy compared to state-of-the-art methods.
Predicting Transcription Factor Binding Sites using Transformer based Capsule Network
Oct 23, 2023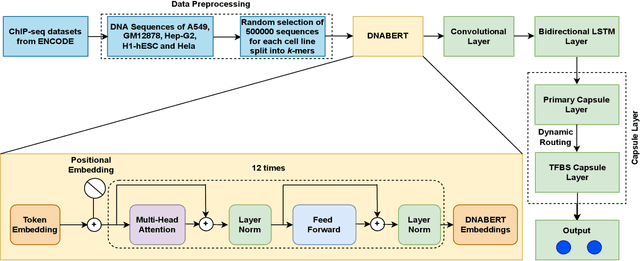
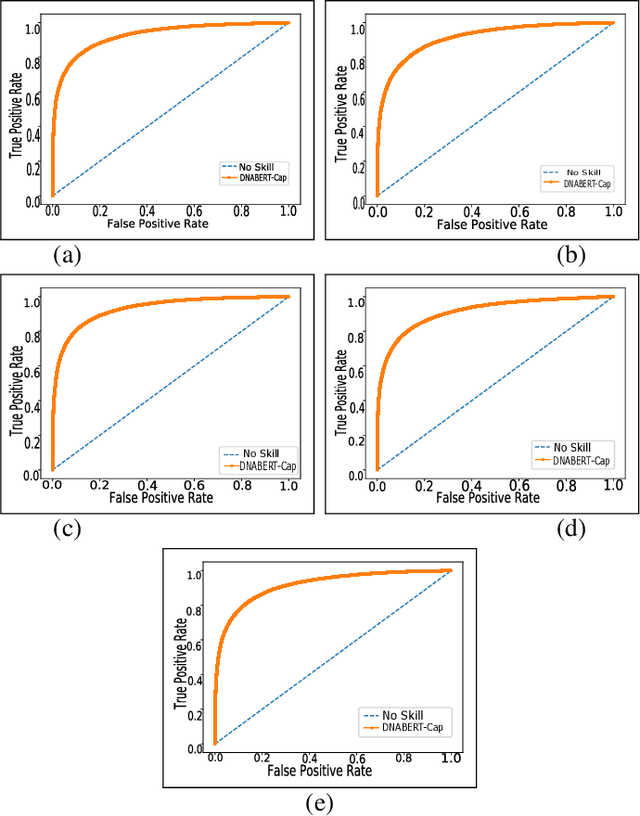
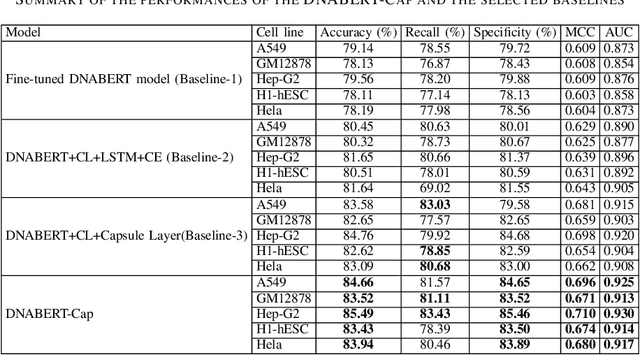
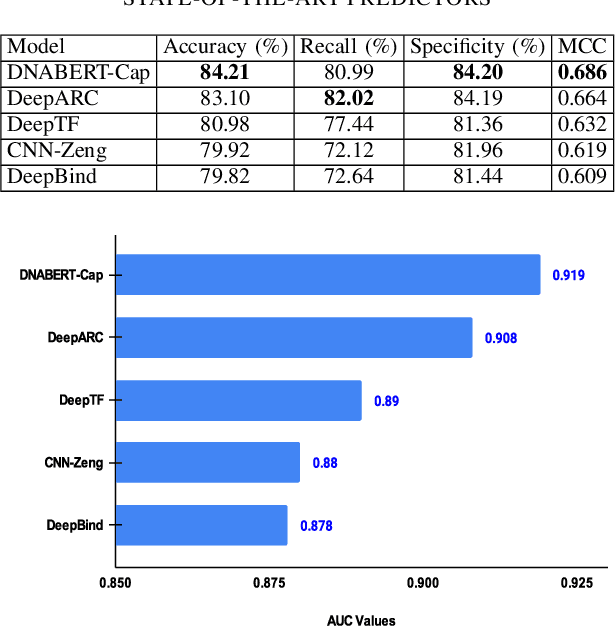
Abstract:Prediction of binding sites for transcription factors is important to understand how they regulate gene expression and how this regulation can be modulated for therapeutic purposes. Although in the past few years there are significant works addressing this issue, there is still space for improvement. In this regard, a transformer based capsule network viz. DNABERT-Cap is proposed in this work to predict transcription factor binding sites mining ChIP-seq datasets. DNABERT-Cap is a bidirectional encoder pre-trained with large number of genomic DNA sequences, empowered with a capsule layer responsible for the final prediction. The proposed model builds a predictor for transcription factor binding sites using the joint optimisation of features encompassing both bidirectional encoder and capsule layer, along with convolutional and bidirectional long-short term memory layers. To evaluate the efficiency of the proposed approach, we use a benchmark ChIP-seq datasets of five cell lines viz. A549, GM12878, Hep-G2, H1-hESC and Hela, available in the ENCODE repository. The results show that the average area under the receiver operating characteristic curve score exceeds 0.91 for all such five cell lines. DNABERT-Cap is also compared with existing state-of-the-art deep learning based predictors viz. DeepARC, DeepTF, CNN-Zeng and DeepBind, and is seen to outperform them.
Hacking Smart Machines with Smarter Ones: How to Extract Meaningful Data from Machine Learning Classifiers
Jun 19, 2013
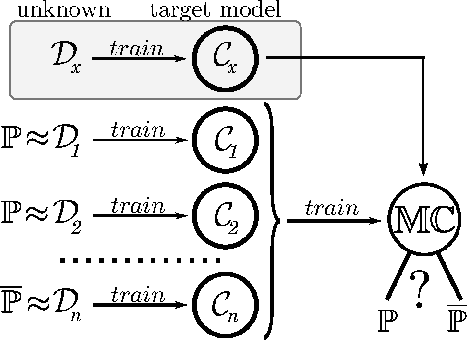
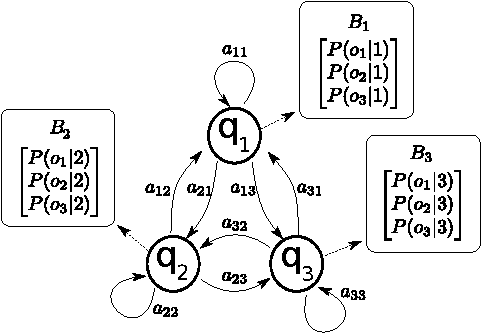
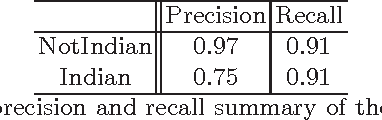
Abstract:Machine Learning (ML) algorithms are used to train computers to perform a variety of complex tasks and improve with experience. Computers learn how to recognize patterns, make unintended decisions, or react to a dynamic environment. Certain trained machines may be more effective than others because they are based on more suitable ML algorithms or because they were trained through superior training sets. Although ML algorithms are known and publicly released, training sets may not be reasonably ascertainable and, indeed, may be guarded as trade secrets. While much research has been performed about the privacy of the elements of training sets, in this paper we focus our attention on ML classifiers and on the statistical information that can be unconsciously or maliciously revealed from them. We show that it is possible to infer unexpected but useful information from ML classifiers. In particular, we build a novel meta-classifier and train it to hack other classifiers, obtaining meaningful information about their training sets. This kind of information leakage can be exploited, for example, by a vendor to build more effective classifiers or to simply acquire trade secrets from a competitor's apparatus, potentially violating its intellectual property rights.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge