Ilias Tagkopoulos
SkillFlow: Efficient Skill and Code Transfer Through Communication in Adapting AI Agents
Apr 08, 2025



Abstract:AI agents are autonomous systems that can execute specific tasks based on predefined programming. Here, we present SkillFlow, a modular, technology-agnostic framework that allows agents to expand their functionality in an ad-hoc fashion by acquiring new skills from their environment or other agents. We present a theoretical model that examines under which conditions this framework would be beneficial, and we then explore SkillFlow's ability to accelerate task completion and lead to lower cumulative costs in a real-world application, namely scheduling agents for calendar events. We demonstrate that within a few iterations, SkillFlow leads to considerable (24.8%, p-value = $6.4\times10^{-3}$) gains in time and cost, especially when the communication cost is high. Finally, we draw analogies from well-studied biological systems and compare this framework to that of lateral gene transfer, a significant process of adaptation and evolution in novel environments.
Interpreting Inflammation Prediction Model via Tag-based Cohort Explanation
Oct 17, 2024Abstract:Machine learning is revolutionizing nutrition science by enabling systems to learn from data and make intelligent decisions. However, the complexity of these models often leads to challenges in understanding their decision-making processes, necessitating the development of explainability techniques to foster trust and increase model transparency. An under-explored type of explanation is cohort explanation, which provides explanations to groups of instances with similar characteristics. Unlike traditional methods that focus on individual explanations or global model behavior, cohort explainability bridges the gap by providing unique insights at an intermediate granularity. We propose a novel framework for identifying cohorts within a dataset based on local feature importance scores, aiming to generate concise descriptions of the clusters via tags. We evaluate our framework on a food-based inflammation prediction model and demonstrated that the framework can generate reliable explanations that match domain knowledge.
Semi-Automated Construction of Food Composition Knowledge Base
Jan 24, 2023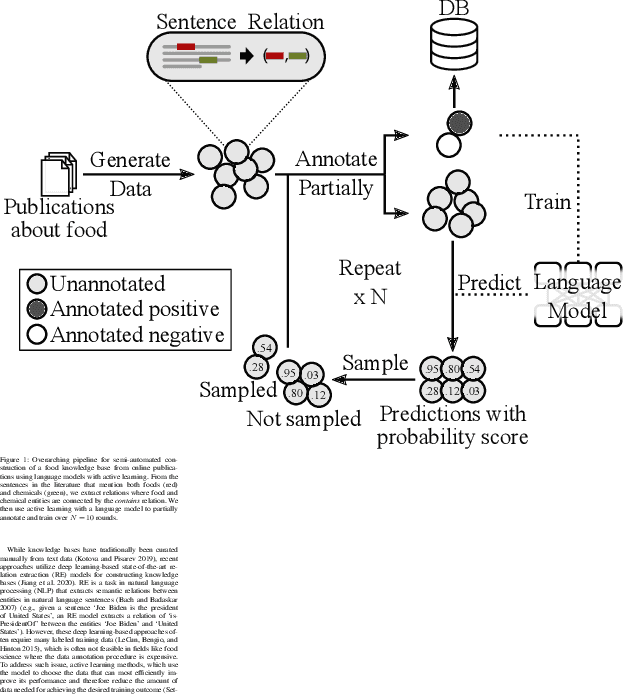
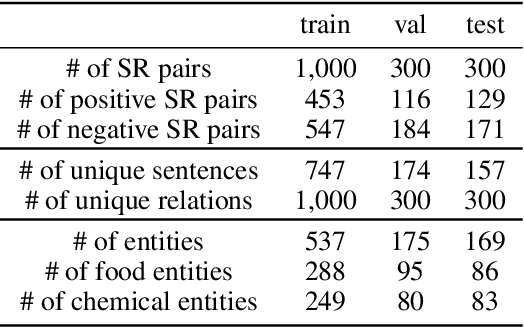
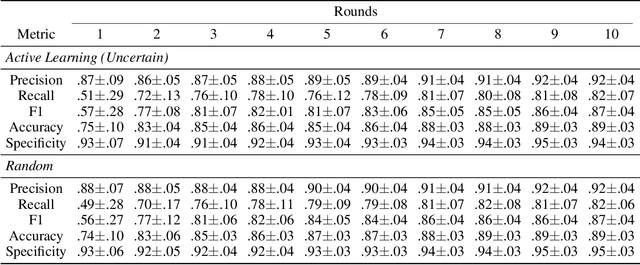
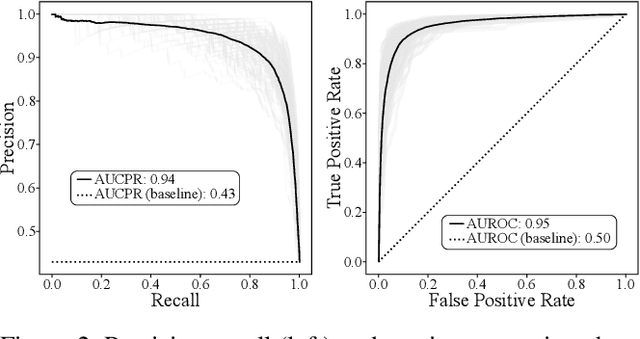
Abstract:A food composition knowledge base, which stores the essential phyto-, micro-, and macro-nutrients of foods is useful for both research and industrial applications. Although many existing knowledge bases attempt to curate such information, they are often limited by time-consuming manual curation processes. Outside of the food science domain, natural language processing methods that utilize pre-trained language models have recently shown promising results for extracting knowledge from unstructured text. In this work, we propose a semi-automated framework for constructing a knowledge base of food composition from the scientific literature available online. To this end, we utilize a pre-trained BioBERT language model in an active learning setup that allows the optimal use of limited training data. Our work demonstrates how human-in-the-loop models are a step toward AI-assisted food systems that scale well to the ever-increasing big data.
KGLM: Integrating Knowledge Graph Structure in Language Models for Link Prediction
Nov 04, 2022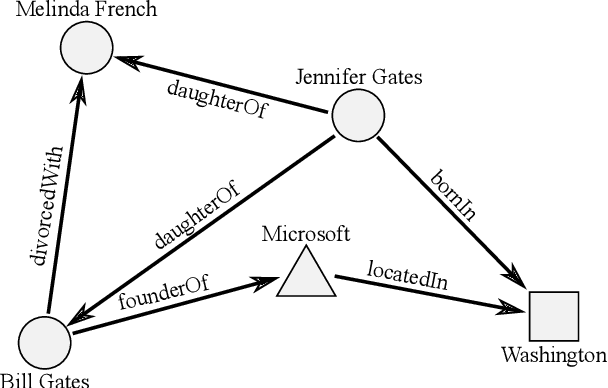
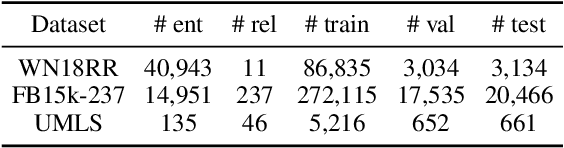
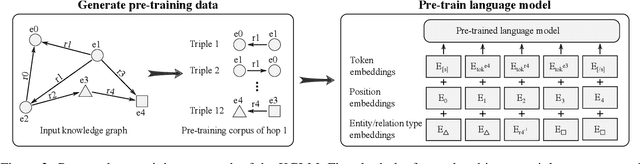
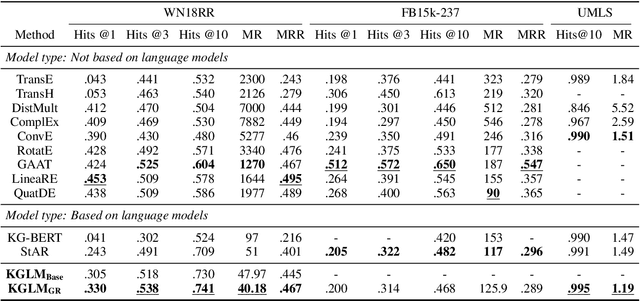
Abstract:The ability of knowledge graphs to represent complex relationships at scale has led to their adoption for various needs including knowledge representation, question-answering, fraud detection, and recommendation systems. Knowledge graphs are often incomplete in the information they represent, necessitating the need for knowledge graph completion tasks, such as link and relation prediction. Pre-trained and fine-tuned language models have shown promise in these tasks although these models ignore the intrinsic information encoded in the knowledge graph, namely the entity and relation types. In this work, we propose the Knowledge Graph Language Model (KGLM) architecture, where we introduce a new entity/relation embedding layer that learns to differentiate distinctive entity and relation types, therefore allowing the model to learn the structure of the knowledge graph. In this work, we show that further pre-training the language models with this additional embedding layer using the triples extracted from the knowledge graph, followed by the standard fine-tuning phase sets a new state-of-the-art performance for the link prediction task on the benchmark datasets.
BWCNN: Blink to Word, a Real-Time Convolutional Neural Network Approach
Jun 01, 2020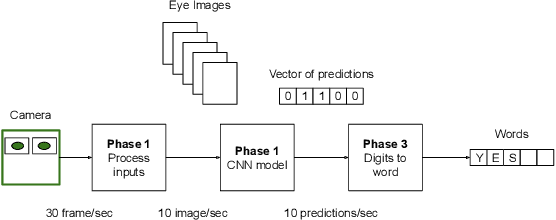

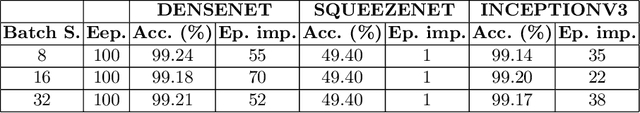
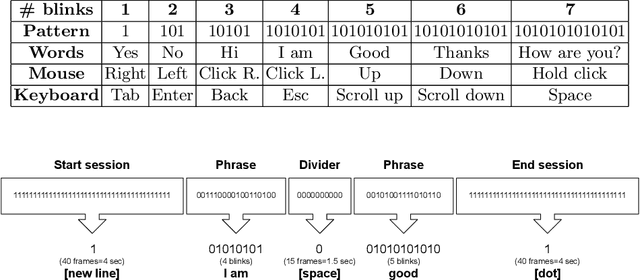
Abstract:Amyotrophic lateral sclerosis (ALS) is a progressive neurodegenerative disease of the brain and the spinal cord, which leads to paralysis of motor functions. Patients retain their ability to blink, which can be used for communication. Here, We present an Artificial Intelligence (AI) system that uses eye-blinks to communicate with the outside world, running on real-time Internet-of-Things (IoT) devices. The system uses a Convolutional Neural Network (CNN) to find the blinking pattern, which is defined as a series of Open and Closed states. Each pattern is mapped to a collection of words that manifest the patient's intent. To investigate the best trade-off between accuracy and latency, we investigated several Convolutional Network architectures, such as ResNet, SqueezeNet, DenseNet, and InceptionV3, and evaluated their performance. We found that the InceptionV3 architecture, after hyper-parameter fine-tuning on the specific task led to the best performance with an accuracy of 99.20% and 94ms latency. This work demonstrates how the latest advances in deep learning architectures can be adapted for clinical systems that ameliorate the patient's quality of life regardless of the point-of-care.
A Distribution Adaptive Framework for Prediction Interval Estimation Using Nominal Variables
Nov 20, 2015



Abstract:Proposed methods for prediction interval estimation so far focus on cases where input variables are numerical. In datasets with solely nominal input variables, we observe records with the exact same input $x^u$, but different real valued outputs due to the inherent noise in the system. Existing prediction interval estimation methods do not use representations that can accurately model such inherent noise in the case of nominal inputs. We propose a new prediction interval estimation method tailored for this type of data, which is prevalent in biology and medicine. We call this method Distribution Adaptive Prediction Interval Estimation given Nominal inputs (DAPIEN) and has four main phases. First, we select a distribution function that can best represent the inherent noise of the system for all unique inputs. Then we infer the parameters $\theta_i$ (e.g. $\theta_i=[mean_i, variance_i]$) of the selected distribution function for all unique input vectors $x^u_i$ and generate a new corresponding training set using pairs of $x^u_i, \theta_i$. III). Then, we train a model to predict $\theta$ given a new $x_u$. Finally, we calculate the prediction interval for a new sample using the inverse of the cumulative distribution function once the parameters $\theta$ is predicted by the trained model. We compared DAPIEN to the commonly used Bootstrap method on three synthetic datasets. Our results show that DAPIEN provides tighter prediction intervals while preserving the requested coverage when compared to Bootstrap. This work can facilitate broader usage of regression methods in medicine and biology where it is necessary to provide tight prediction intervals while preserving coverage when input variables are nominal.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge