Huan Minh Luu
Biomedical image analysis competitions: The state of current participation practice
Dec 16, 2022Abstract:The number of international benchmarking competitions is steadily increasing in various fields of machine learning (ML) research and practice. So far, however, little is known about the common practice as well as bottlenecks faced by the community in tackling the research questions posed. To shed light on the status quo of algorithm development in the specific field of biomedical imaging analysis, we designed an international survey that was issued to all participants of challenges conducted in conjunction with the IEEE ISBI 2021 and MICCAI 2021 conferences (80 competitions in total). The survey covered participants' expertise and working environments, their chosen strategies, as well as algorithm characteristics. A median of 72% challenge participants took part in the survey. According to our results, knowledge exchange was the primary incentive (70%) for participation, while the reception of prize money played only a minor role (16%). While a median of 80 working hours was spent on method development, a large portion of participants stated that they did not have enough time for method development (32%). 25% perceived the infrastructure to be a bottleneck. Overall, 94% of all solutions were deep learning-based. Of these, 84% were based on standard architectures. 43% of the respondents reported that the data samples (e.g., images) were too large to be processed at once. This was most commonly addressed by patch-based training (69%), downsampling (37%), and solving 3D analysis tasks as a series of 2D tasks. K-fold cross-validation on the training set was performed by only 37% of the participants and only 50% of the participants performed ensembling based on multiple identical models (61%) or heterogeneous models (39%). 48% of the respondents applied postprocessing steps.
Extending nn-UNet for brain tumor segmentation
Dec 09, 2021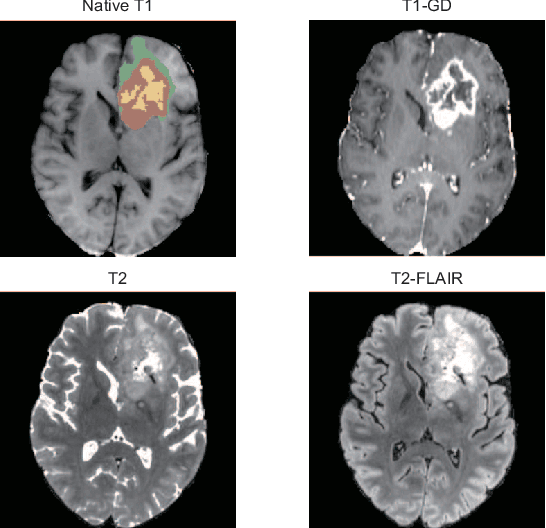
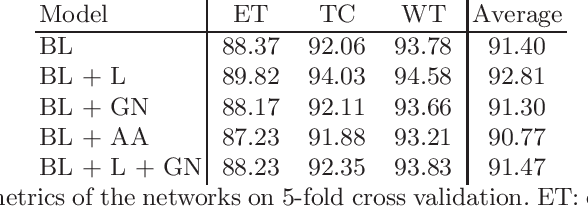
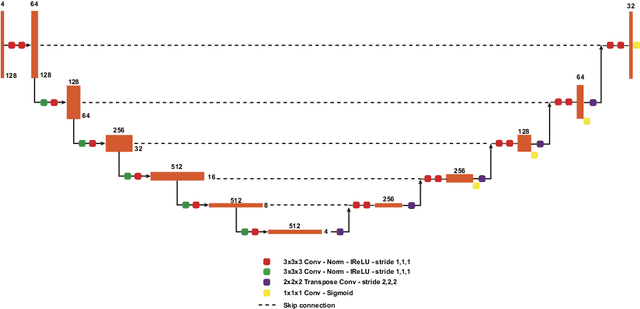
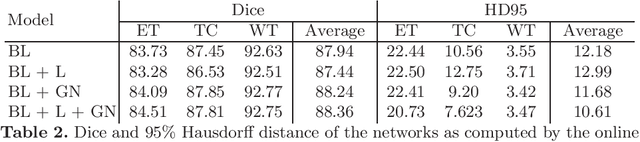
Abstract:Brain tumor segmentation is essential for the diagnosis and prognosis of patients with gliomas. The brain tumor segmentation challenge has continued to provide a great source of data to develop automatic algorithms to perform the task. This paper describes our contribution to the 2021 competition. We developed our methods based on nn-UNet, the winning entry of last year competition. We experimented with several modifications, including using a larger network, replacing batch normalization with group normalization, and utilizing axial attention in the decoder. Internal 5-fold cross validation as well as online evaluation from the organizers showed the effectiveness of our approach, with minor improvement in quantitative metrics when compared to the baseline. The proposed models won first place in the final ranking on unseen test data. The codes, pretrained weights, and docker image for the winning submission are publicly available at https://github.com/rixez/Brats21_KAIST_MRI_Lab
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge