Hrithwik Shalu
Uncertainty Quantification in Detecting Choroidal Metastases on MRI via Evolutionary Strategies
Apr 12, 2024Abstract:Uncertainty quantification plays a vital role in facilitating the practical implementation of AI in radiology by addressing growing concerns around trustworthiness. Given the challenges associated with acquiring large, annotated datasets in this field, there is a need for methods that enable uncertainty quantification in small data AI approaches tailored to radiology images. In this study, we focused on uncertainty quantification within the context of the small data evolutionary strategies-based technique of deep neuroevolution (DNE). Specifically, we employed DNE to train a simple Convolutional Neural Network (CNN) with MRI images of the eyes for binary classification. The goal was to distinguish between normal eyes and those with metastatic tumors called choroidal metastases. The training set comprised 18 images with choroidal metastases and 18 without tumors, while the testing set contained a tumor-to-normal ratio of 15:15. We trained CNN model weights via DNE for approximately 40,000 episodes, ultimately reaching a convergence of 100% accuracy on the training set. We saved all models that achieved maximal training set accuracy. Then, by applying these models to the testing set, we established an ensemble method for uncertainty quantification.The saved set of models produced distributions for each testing set image between the two classes of normal and tumor-containing. The relative frequencies permitted uncertainty quantification of model predictions. Intriguingly, we found that subjective features appreciated by human radiologists explained images for which uncertainty was high, highlighting the significance of uncertainty quantification in AI-driven radiological analyses.
Kinematics Modeling of Peroxy Free Radicals: A Deep Reinforcement Learning Approach
Apr 12, 2024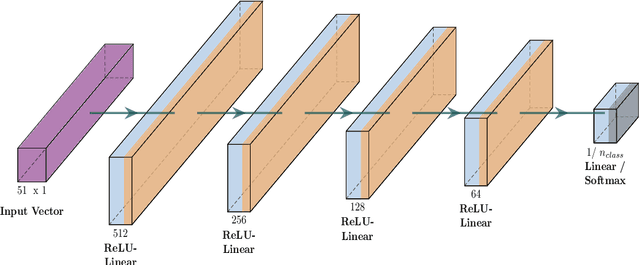
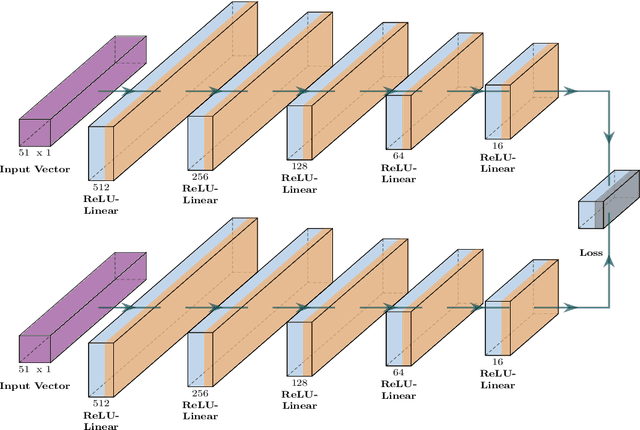
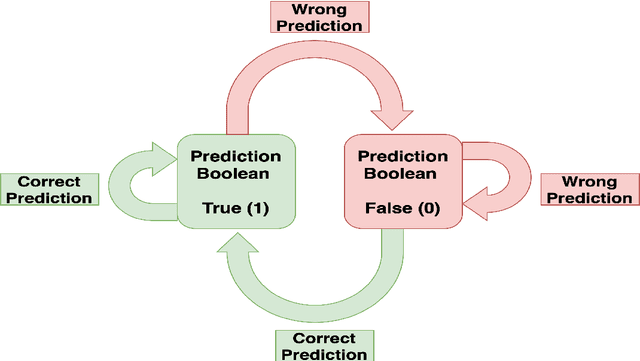
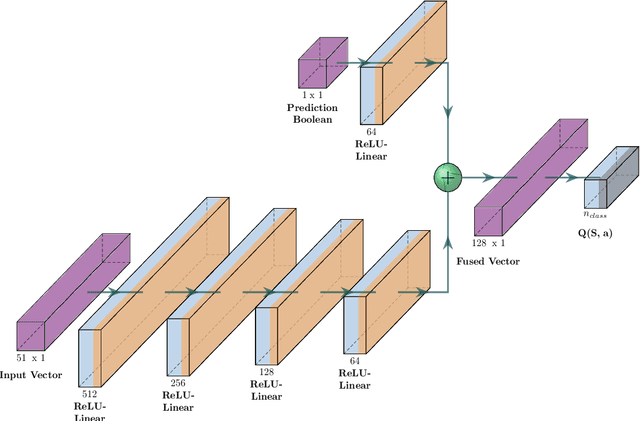
Abstract:Tropospheric ozone, known as a concerning air pollutant, has been associated with health issues including asthma, bronchitis, and impaired lung function. The rates at which peroxy radicals react with NO play a critical role in the overall formation and depletion of tropospheric ozone. However, obtaining comprehensive kinetic data for these reactions remains challenging. Traditional approaches to determine rate constants are costly and technically intricate. Fortunately, the emergence of machine learning-based models offers a less resource and time-intensive alternative for acquiring kinetics information. In this study, we leveraged deep reinforcement learning to predict ranges of rate constants (\textit{k}) with exceptional accuracy, achieving a testing set accuracy of 100%. To analyze reactivity trends based on the molecular structure of peroxy radicals, we employed 51 global descriptors as input parameters. These descriptors were derived from optimized minimum energy geometries of peroxy radicals using the quantum composite G3B3 method. Through the application of Integrated Gradients (IGs), we gained valuable insights into the significance of the various descriptors in relation to reaction rates. We successfully validated and contextualized our findings by conducting cross-comparisons with established trends in the existing literature. These results establish a solid foundation for pioneering advancements in chemistry, where computer analysis serves as an inspirational source driving innovation.
Deep neuroevolution to predict primary brain tumor grade from functional MRI adjacency matrices
Nov 26, 2022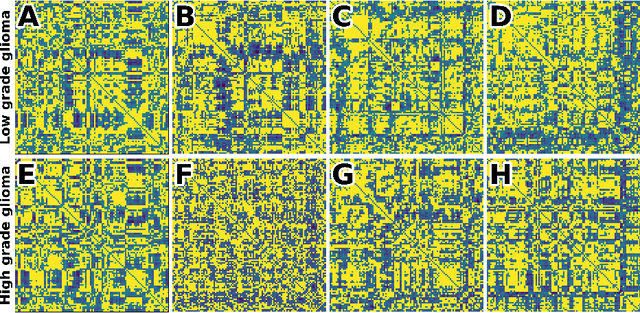
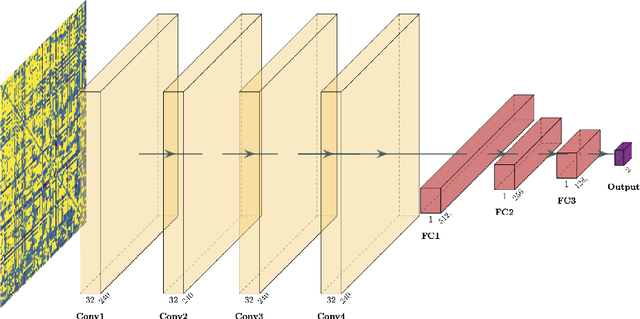
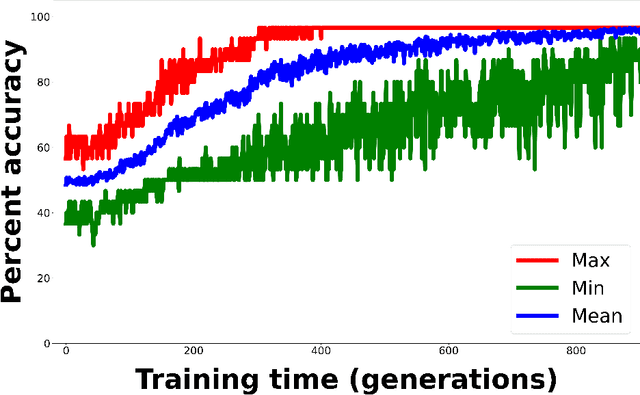
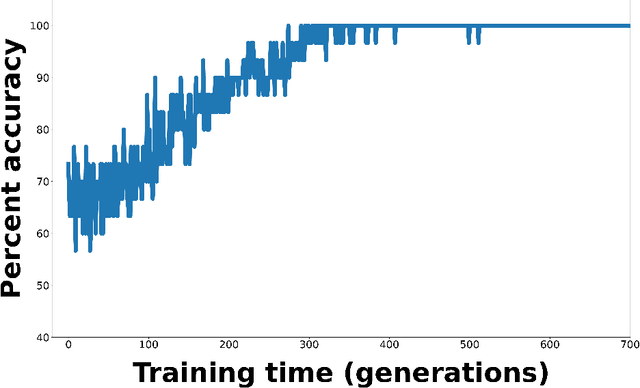
Abstract:Whereas MRI produces anatomic information about the brain, functional MRI (fMRI) tells us about neural activity within the brain, including how various regions communicate with each other. The full chorus of conversations within the brain is summarized elegantly in the adjacency matrix. Although information-rich, adjacency matrices typically provide little in the way of intuition. Whereas trained radiologists viewing anatomic MRI can readily distinguish between different kinds of brain cancer, a similar determination using adjacency matrices would exceed any expert's grasp. Artificial intelligence (AI) in radiology usually analyzes anatomic imaging, providing assistance to radiologists. For non-intuitive data types such as adjacency matrices, AI moves beyond the role of helpful assistant, emerging as indispensible. We sought here to show that AI can learn to discern between two important brain tumor types, high-grade glioma (HGG) and low-grade glioma (LGG), based on adjacency matrices. We trained a convolutional neural networks (CNN) with the method of deep neuroevolution (DNE), because of the latter's recent promising results; DNE has produced remarkably accurate CNNs even when relying on small and noisy training sets, or performing nuanced tasks. After training on just 30 adjacency matrices, our CNN could tell HGG apart from LGG with perfect testing set accuracy. Saliency maps revealed that the network learned highly sophisticated and complex features to achieve its success. Hence, we have shown that it is possible for AI to recognize brain tumor type from functional connectivity. In future work, we will apply DNE to other noisy and somewhat cryptic forms of medical data, including further explorations with fMRI.
Deep neuroevolution for limited, heterogeneous data: proof-of-concept application to Neuroblastoma brain metastasis using a small virtual pooled image collection
Nov 26, 2022



Abstract:Artificial intelligence (AI) in radiology has made great strides in recent years, but many hurdles remain. Overfitting and lack of generalizability represent important ongoing challenges hindering accurate and dependable clinical deployment. If AI algorithms can avoid overfitting and achieve true generalizability, they can go from the research realm to the forefront of clinical work. Recently, small data AI approaches such as deep neuroevolution (DNE) have avoided overfitting small training sets. We seek to address both overfitting and generalizability by applying DNE to a virtually pooled data set consisting of images from various institutions. Our use case is classifying neuroblastoma brain metastases on MRI. Neuroblastoma is well-suited for our goals because it is a rare cancer. Hence, studying this pediatric disease requires a small data approach. As a tertiary care center, the neuroblastoma images in our local Picture Archiving and Communication System (PACS) are largely from outside institutions. These multi-institutional images provide a heterogeneous data set that can simulate real world clinical deployment. As in prior DNE work, we used a small training set, consisting of 30 normal and 30 metastasis-containing post-contrast MRI brain scans, with 37% outside images. The testing set was enriched with 83% outside images. DNE converged to a testing set accuracy of 97%. Hence, the algorithm was able to predict image class with near-perfect accuracy on a testing set that simulates real-world data. Hence, the work described here represents a considerable contribution toward clinically feasible AI.
Deep reinforcement learning for fMRI prediction of Autism Spectrum Disorder
Jun 17, 2022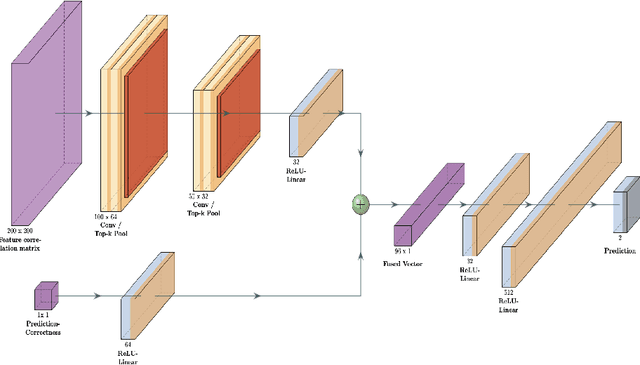
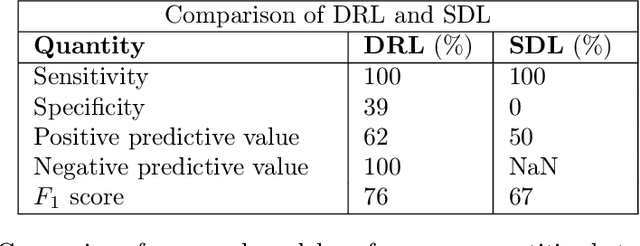
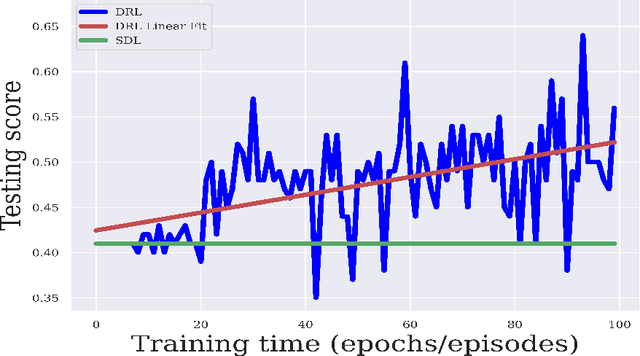
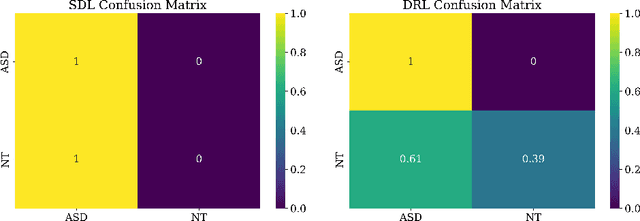
Abstract:Purpose : Because functional MRI (fMRI) data sets are in general small, we sought a data efficient approach to resting state fMRI classification of autism spectrum disorder (ASD) versus neurotypical (NT) controls. We hypothesized that a Deep Reinforcement Learning (DRL) classifier could learn effectively on a small fMRI training set. Methods : We trained a Deep Reinforcement Learning (DRL) classifier on 100 graph-label pairs from the Autism Brain Imaging Data Exchange (ABIDE) database. For comparison, we trained a Supervised Deep Learning (SDL) classifier on the same training set. Results : DRL significantly outperformed SDL, with a p-value of 2.4 x 10^(-7). DRL achieved superior results for a variety of classifier performance metrics, including an F1 score of 76, versus 67 for SDL. Whereas SDL quickly overfit the training data, DRL learned in a progressive manner that generalised to the separate testing set. Conclusion : DRL can learn to classify ASD versus NT in a data efficient manner, doing so for a small training set. Future work will involve optimizing the neural network for data efficiency and applying the approach to other fMRI data sets, namely for brain cancer patients.
Direct evaluation of progression or regression of disease burden in brain metastatic disease with Deep Neuroevolution
Mar 24, 2022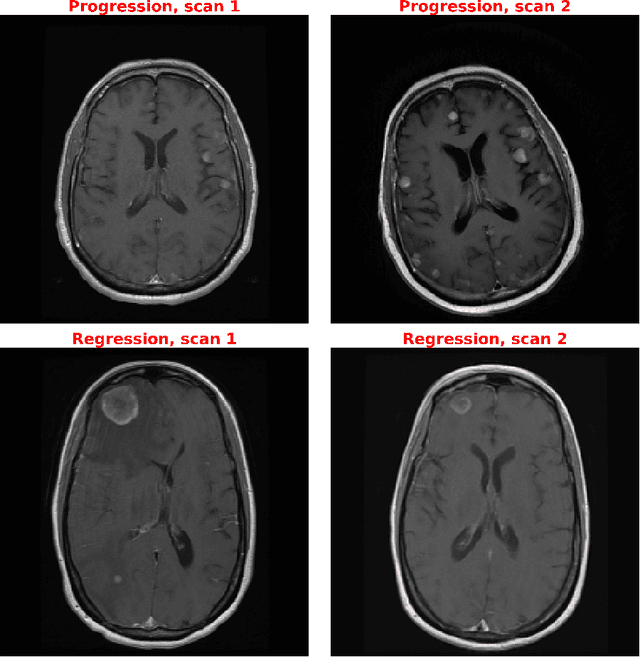
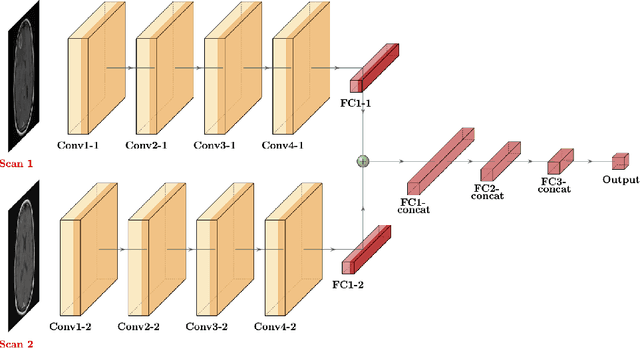
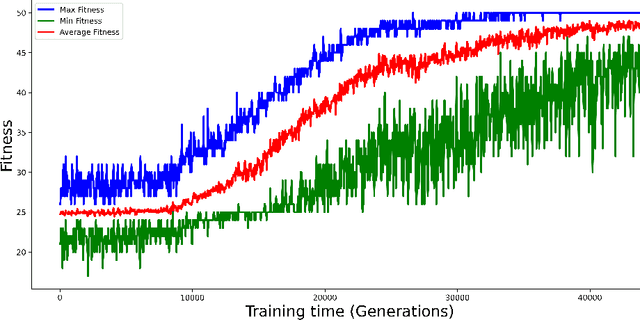
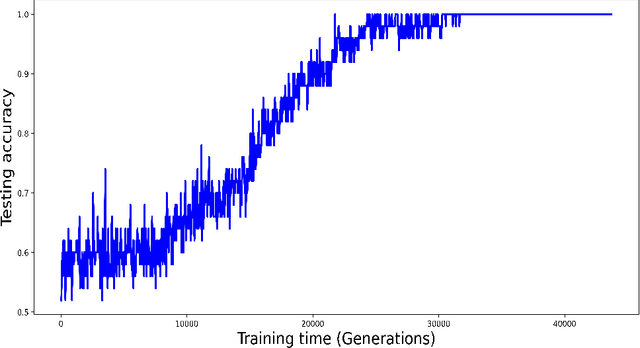
Abstract:Purpose: A core component of advancing cancer treatment research is assessing response to therapy. Doing so by hand, for example as per RECIST or RANO criteria, is tedious, time-consuming, and can miss important tumor response information; most notably, they exclude non-target lesions. We wish to assess change in a holistic fashion that includes all lesions, obtaining simple, informative, and automated assessments of tumor progression or regression. Due to often low patient enrolments in clinical trials, we wish to make response assessments with small training sets. Deep neuroevolution (DNE) can produce radiology artificial intelligence (AI) that performs well on small training sets. Here we use DNE for function approximation that predicts progression versus regression of metastatic brain disease. Methods: We analyzed 50 pairs of MRI contrast-enhanced images as our training set. Half of these pairs, separated in time, qualified as disease progression, while the other 25 images constituted regression. We trained the parameters of a relatively small CNN via mutations that consisted of random CNN weight adjustments and mutation fitness. We then incorporated the best mutations into the next generations CNN, repeating this process for approximately 50,000 generations. We applied the CNNs to our training set, as well as a separate testing set with the same class balance of 25 progression and 25 regression images. Results: DNE achieved monotonic convergence to 100% training set accuracy. DNE also converged monotonically to 100% testing set accuracy. Conclusion: DNE can accurately classify brain-metastatic disease progression versus regression. Future work will extend the input from 2D image slices to full 3D volumes, and include the category of no change. We believe that an approach such as our could ultimately provide a useful adjunct to RANO/RECIST assessment.
Deep Neuroevolution Squeezes More out of Small Neural Networks and Small Training Sets: Sample Application to MRI Brain Sequence Classification
Dec 24, 2021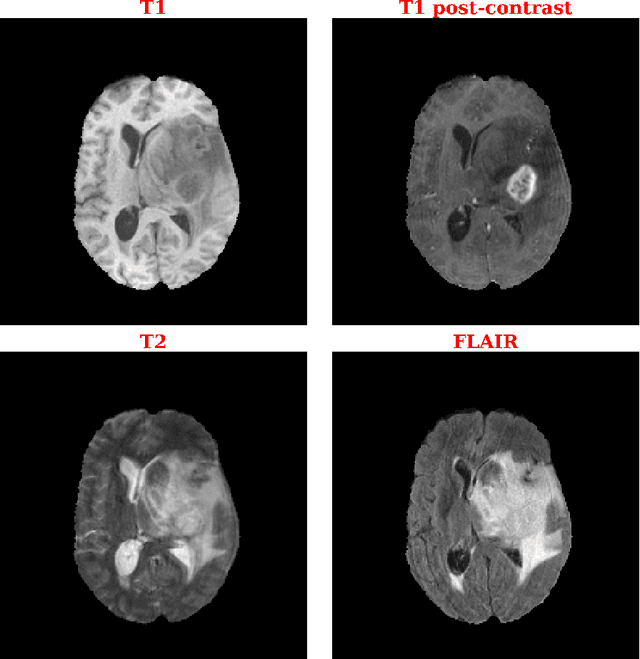
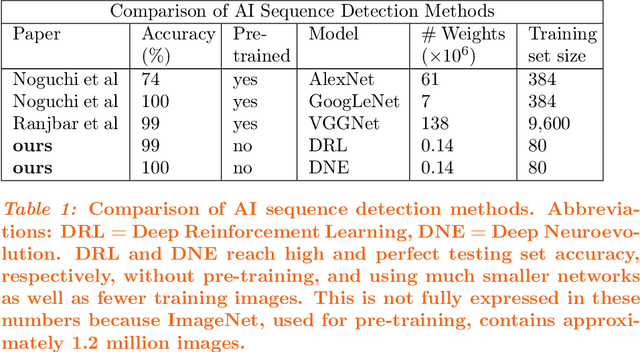
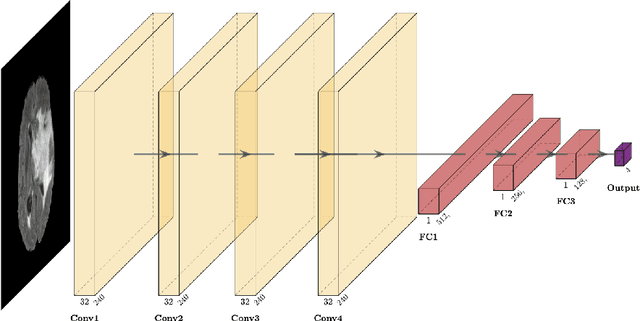
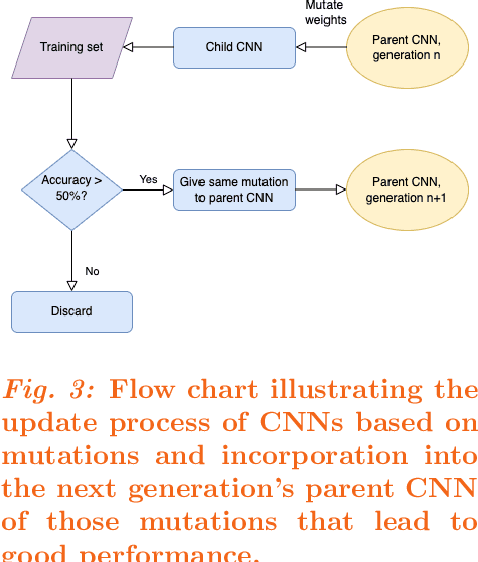
Abstract:Purpose: Deep Neuroevolution (DNE) holds the promise of providing radiology artificial intelligence (AI) that performs well with small neural networks and small training sets. We seek to realize this potential via a proof-of-principle application to MRI brain sequence classification. Methods: We analyzed a training set of 20 patients, each with four sequences/weightings: T1, T1 post-contrast, T2, and T2-FLAIR. We trained the parameters of a relatively small convolutional neural network (CNN) as follows: First, we randomly mutated the CNN weights. We then measured the CNN training set accuracy, using the latter as the fitness evaluation metric. The fittest child CNNs were identified. We incorporated their mutations into the parent CNN. This selectively mutated parent became the next generation's parent CNN. We repeated this process for approximately 50,000 generations. Results: DNE achieved monotonic convergence to 100% training set accuracy. DNE also converged monotonically to 100% testing set accuracy. Conclusions: DNE can achieve perfect accuracy with small training sets and small CNNs. Particularly when combined with Deep Reinforcement Learning, DNE may provide a path forward in the quest to make radiology AI more human-like in its ability to learn. DNE may very well turn out to be a key component of the much-anticipated meta-learning regime of radiology AI algorithms that can adapt to new tasks and new image types, similar to human radiologists.
Efficient Feature Representations for Cricket Data Analysis using Deep Learning based Multi-Modal Fusion Model
Aug 16, 2021
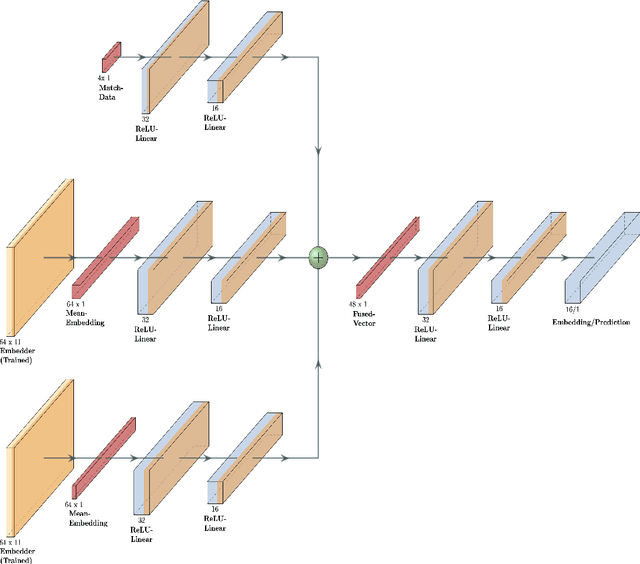
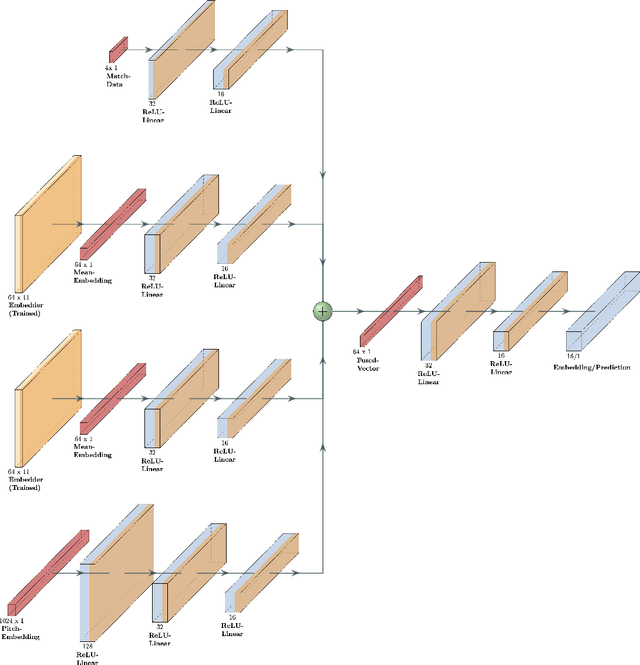

Abstract:Data analysis has become a necessity in the modern era of cricket. Everything from effective team management to match win predictions use some form of analytics. Meaningful data representations are necessary for efficient analysis of data. In this study we investigate the use of adaptive (learnable) embeddings to represent inter-related features (such as players, teams, etc). The data used for this study is collected from a classical T20 tournament IPL (Indian Premier League). To naturally facilitate the learning of meaningful representations of features for accurate data analysis, we formulate a deep representation learning framework which jointly learns a custom set of embeddings (which represents our features of interest) through the minimization of a contrastive loss. We base our objective on a set of classes obtained as a result of hierarchical clustering on the overall run rate of an innings. It's been assessed that the framework ensures greater generality in the obtained embeddings, on top of which a task based analysis of overall run rate prediction was done to show the reliability of the framework.
Deep reinforcement learning with automated label extraction from clinical reports accurately classifies 3D MRI brain volumes
Jun 17, 2021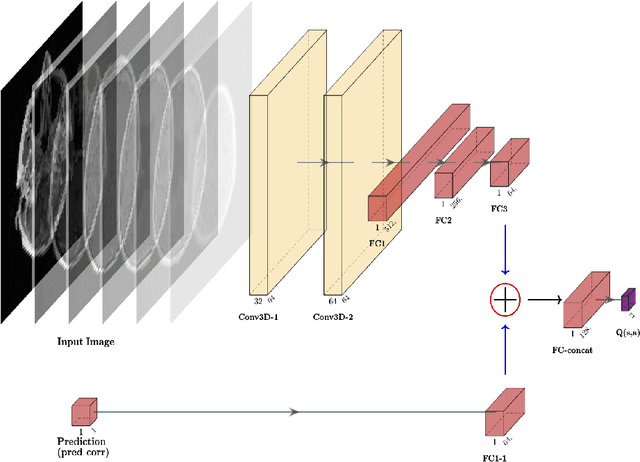
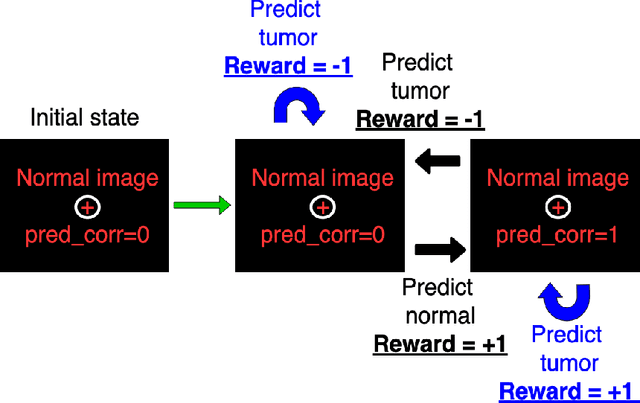
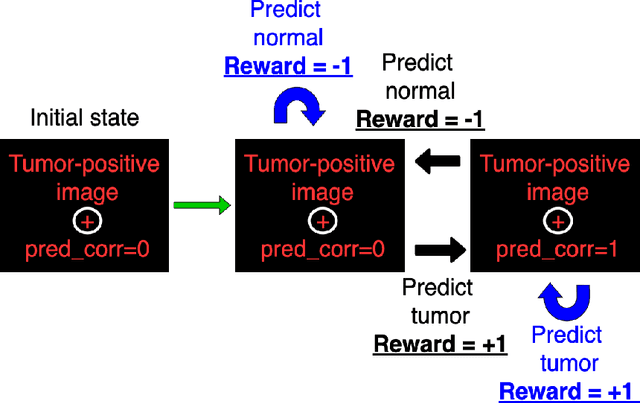
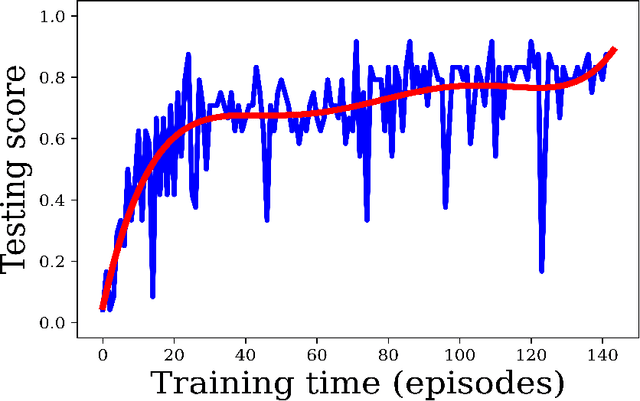
Abstract:Purpose: Image classification is perhaps the most fundamental task in imaging AI. However, labeling images is time-consuming and tedious. We have recently demonstrated that reinforcement learning (RL) can classify 2D slices of MRI brain images with high accuracy. Here we make two important steps toward speeding image classification: Firstly, we automatically extract class labels from the clinical reports. Secondly, we extend our prior 2D classification work to fully 3D image volumes from our institution. Hence, we proceed as follows: in Part 1, we extract labels from reports automatically using the SBERT natural language processing approach. Then, in Part 2, we use these labels with RL to train a classification Deep-Q Network (DQN) for 3D image volumes. Methods: For Part 1, we trained SBERT with 90 radiology report impressions. We then used the trained SBERT to predict class labels for use in Part 2. In Part 2, we applied multi-step image classification to allow for combined Deep-Q learning using 3D convolutions and TD(0) Q learning. We trained on a set of 90 images. We tested on a separate set of 61 images, again using the classes predicted from patient reports by the trained SBERT in Part 1. For comparison, we also trained and tested a supervised deep learning classification network on the same set of training and testing images using the same labels. Results: Part 1: Upon training with the corpus of radiology reports, the SBERT model had 100% accuracy for both normal and metastasis-containing scans. Part 2: Then, using these labels, whereas the supervised approach quickly overfit the training data and as expected performed poorly on the testing set (66% accuracy, just over random guessing), the reinforcement learning approach achieved an accuracy of 92%. The results were found to be statistically significant, with a p-value of 3.1 x 10^-5.
Deep Neural Network Based Differential Equation Solver for HIV Enzyme Kinetics
Feb 16, 2021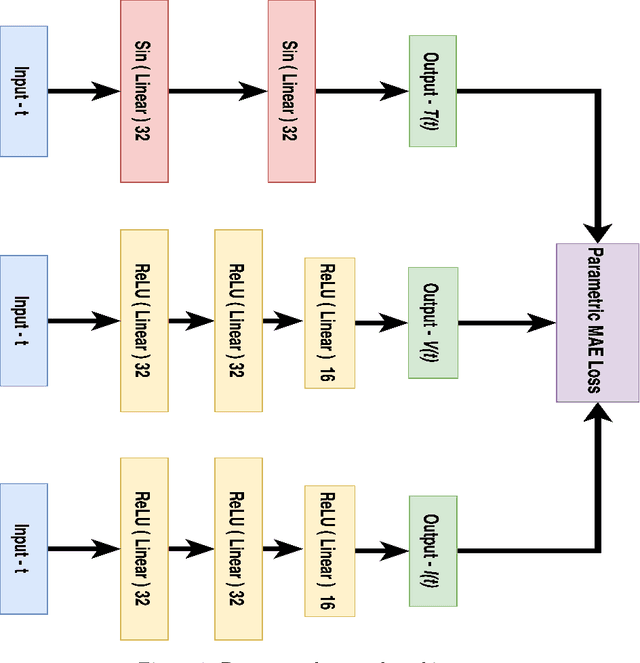
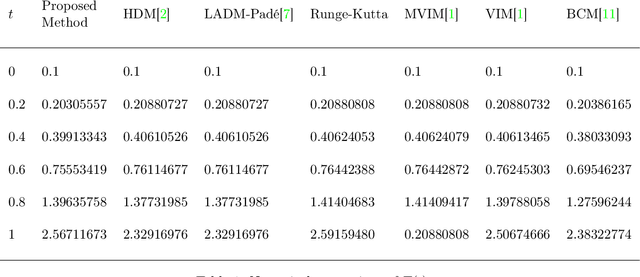
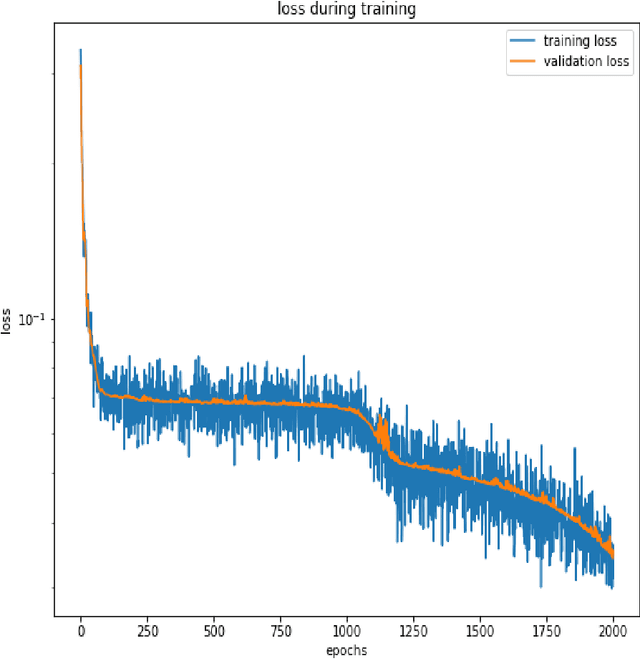
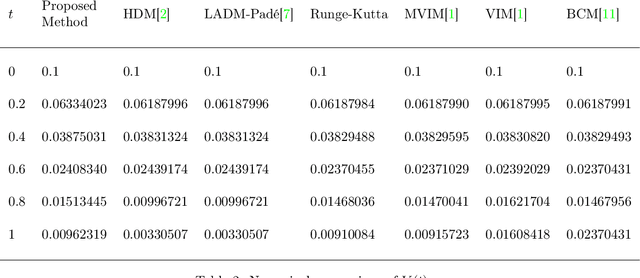
Abstract:Purpose: We seek to use neural networks (NNs) to solve a well-known system of differential equations describing the balance between T cells and HIV viral burden. Materials and Methods: In this paper, we employ a 3-input parallel NN to approximate solutions for the system of first-order ordinary differential equations describing the above biochemical relationship. Results: The numerical results obtained by the NN are very similar to a host of numerical approximations from the literature. Conclusion: We have demonstrated use of NN integration of a well-known and medically important system of first order coupled ordinary differential equations. Our trial-and-error approach counteracts the system's inherent scale imbalance. However, it highlights the need to address scale imbalance more substantively in future work. Doing so will allow more automated solutions to larger systems of equations, which could describe increasingly complex and biologically interesting systems.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge