Andrei Holodny
Uncertainty Quantification in Detecting Choroidal Metastases on MRI via Evolutionary Strategies
Apr 12, 2024Abstract:Uncertainty quantification plays a vital role in facilitating the practical implementation of AI in radiology by addressing growing concerns around trustworthiness. Given the challenges associated with acquiring large, annotated datasets in this field, there is a need for methods that enable uncertainty quantification in small data AI approaches tailored to radiology images. In this study, we focused on uncertainty quantification within the context of the small data evolutionary strategies-based technique of deep neuroevolution (DNE). Specifically, we employed DNE to train a simple Convolutional Neural Network (CNN) with MRI images of the eyes for binary classification. The goal was to distinguish between normal eyes and those with metastatic tumors called choroidal metastases. The training set comprised 18 images with choroidal metastases and 18 without tumors, while the testing set contained a tumor-to-normal ratio of 15:15. We trained CNN model weights via DNE for approximately 40,000 episodes, ultimately reaching a convergence of 100% accuracy on the training set. We saved all models that achieved maximal training set accuracy. Then, by applying these models to the testing set, we established an ensemble method for uncertainty quantification.The saved set of models produced distributions for each testing set image between the two classes of normal and tumor-containing. The relative frequencies permitted uncertainty quantification of model predictions. Intriguingly, we found that subjective features appreciated by human radiologists explained images for which uncertainty was high, highlighting the significance of uncertainty quantification in AI-driven radiological analyses.
Deep neuroevolution to predict primary brain tumor grade from functional MRI adjacency matrices
Nov 26, 2022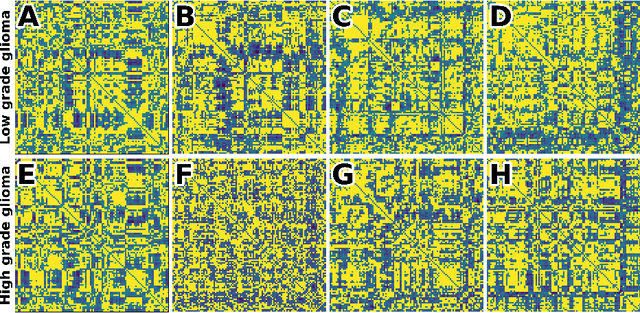
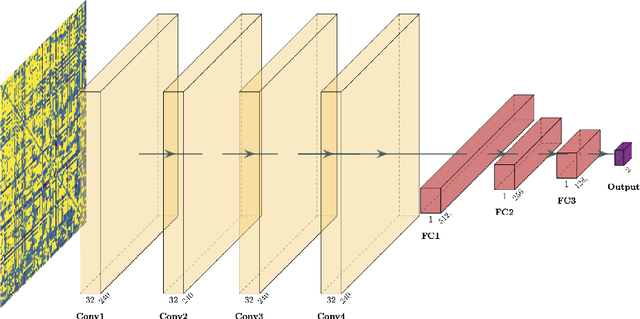
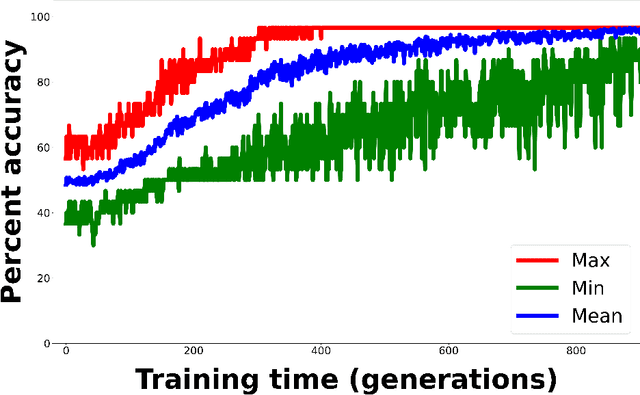
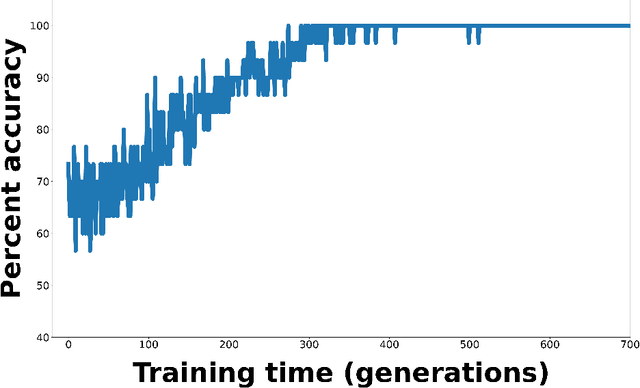
Abstract:Whereas MRI produces anatomic information about the brain, functional MRI (fMRI) tells us about neural activity within the brain, including how various regions communicate with each other. The full chorus of conversations within the brain is summarized elegantly in the adjacency matrix. Although information-rich, adjacency matrices typically provide little in the way of intuition. Whereas trained radiologists viewing anatomic MRI can readily distinguish between different kinds of brain cancer, a similar determination using adjacency matrices would exceed any expert's grasp. Artificial intelligence (AI) in radiology usually analyzes anatomic imaging, providing assistance to radiologists. For non-intuitive data types such as adjacency matrices, AI moves beyond the role of helpful assistant, emerging as indispensible. We sought here to show that AI can learn to discern between two important brain tumor types, high-grade glioma (HGG) and low-grade glioma (LGG), based on adjacency matrices. We trained a convolutional neural networks (CNN) with the method of deep neuroevolution (DNE), because of the latter's recent promising results; DNE has produced remarkably accurate CNNs even when relying on small and noisy training sets, or performing nuanced tasks. After training on just 30 adjacency matrices, our CNN could tell HGG apart from LGG with perfect testing set accuracy. Saliency maps revealed that the network learned highly sophisticated and complex features to achieve its success. Hence, we have shown that it is possible for AI to recognize brain tumor type from functional connectivity. In future work, we will apply DNE to other noisy and somewhat cryptic forms of medical data, including further explorations with fMRI.
Deep reinforcement learning for fMRI prediction of Autism Spectrum Disorder
Jun 17, 2022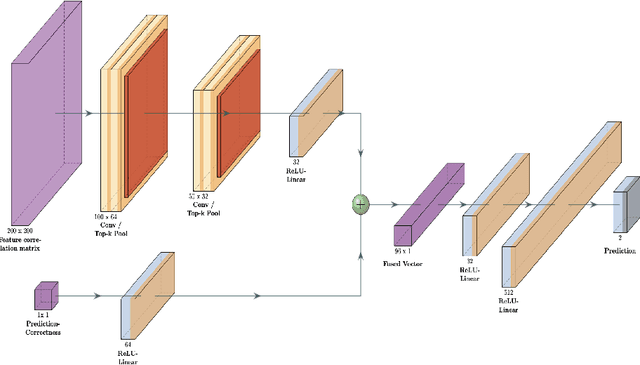
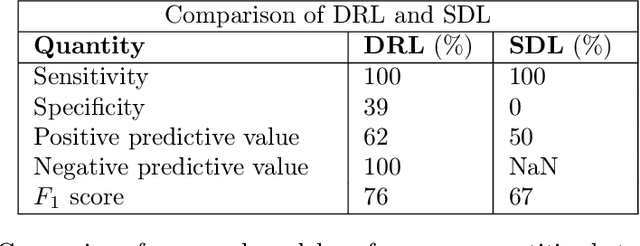
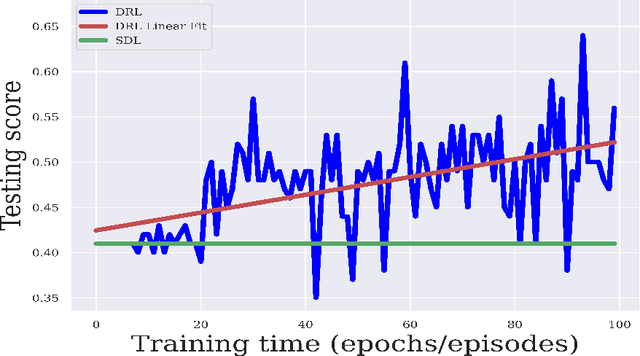
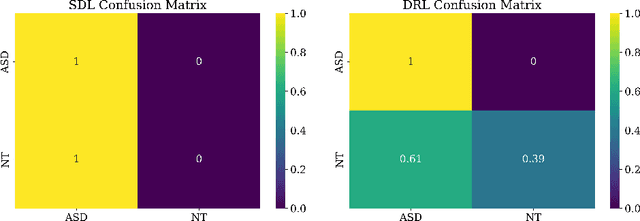
Abstract:Purpose : Because functional MRI (fMRI) data sets are in general small, we sought a data efficient approach to resting state fMRI classification of autism spectrum disorder (ASD) versus neurotypical (NT) controls. We hypothesized that a Deep Reinforcement Learning (DRL) classifier could learn effectively on a small fMRI training set. Methods : We trained a Deep Reinforcement Learning (DRL) classifier on 100 graph-label pairs from the Autism Brain Imaging Data Exchange (ABIDE) database. For comparison, we trained a Supervised Deep Learning (SDL) classifier on the same training set. Results : DRL significantly outperformed SDL, with a p-value of 2.4 x 10^(-7). DRL achieved superior results for a variety of classifier performance metrics, including an F1 score of 76, versus 67 for SDL. Whereas SDL quickly overfit the training data, DRL learned in a progressive manner that generalised to the separate testing set. Conclusion : DRL can learn to classify ASD versus NT in a data efficient manner, doing so for a small training set. Future work will involve optimizing the neural network for data efficiency and applying the approach to other fMRI data sets, namely for brain cancer patients.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge